De novo design of miniprotein antagonists of cytokine storm inducers.
Huang, B., Coventry, B., Borowska, M.T., Arhontoulis, D.C., Exposit, M., Abedi, M., Jude, K.M., Halabiya, S.F., Allen, A., Cordray, C., Goreshnik, I., Ahlrichs, M., Chan, S., Tunggal, H., DeWitt, M., Hyams, N., Carter, L., Stewart, L., Fuller, D.H., Mei, Y., Garcia, K.C., Baker, D.(2024) Nat Commun 15: 7064-7064
- PubMed: 39152100
- DOI: https://doi.org/10.1038/s41467-024-50919-4
- Primary Citation of Related Structures:
8UOS, 8UPA, 8UPB - PubMed Abstract:
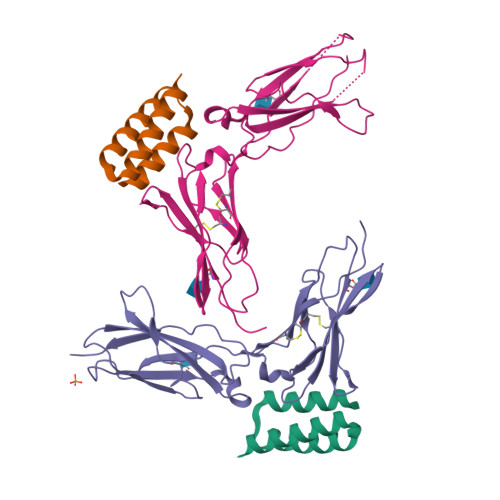
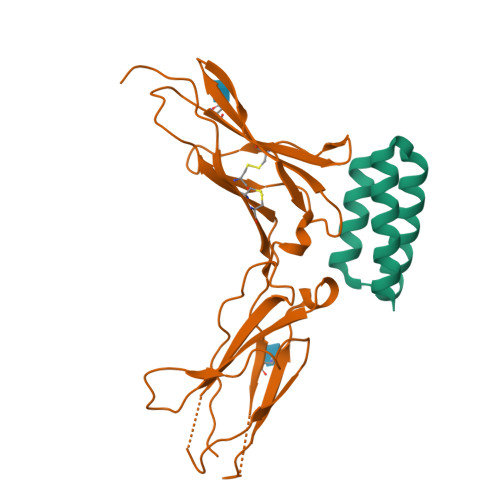
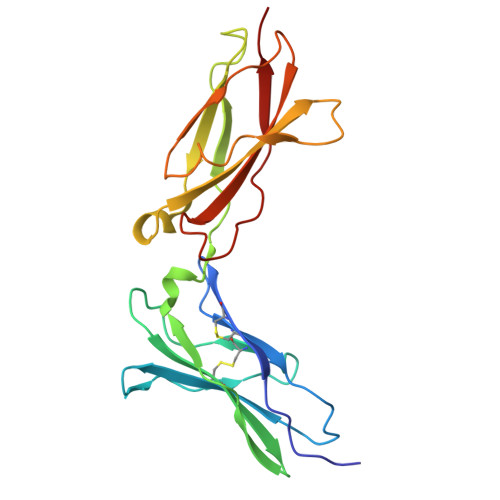
Cytokine release syndrome (CRS), commonly known as cytokine storm, is an acute systemic inflammatory response that is a significant global health threat. Interleukin-6 (IL-6) and interleukin-1 (IL-1) are key pro-inflammatory cytokines involved in CRS and are hence critical therapeutic targets. Current antagonists, such as tocilizumab and anakinra, target IL-6R/IL-1R but have limitations due to their long half-life and systemic anti-inflammatory effects, making them less suitable for acute or localized treatments. Here we present the de novo design of small protein antagonists that prevent IL-1 and IL-6 from interacting with their receptors to activate signaling. The designed proteins bind to the IL-6R, GP130 (an IL-6 co-receptor), and IL-1R1 receptor subunits with binding affinities in the picomolar to low-nanomolar range. X-ray crystallography studies reveal that the structures of these antagonists closely match their computational design models. In a human cardiac organoid disease model, the IL-1R antagonists demonstrated protective effects against inflammation and cardiac damage induced by IL-1β. These minibinders show promise for administration via subcutaneous injection or intranasal/inhaled routes to mitigate acute cytokine storm effects.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, WA, USA.