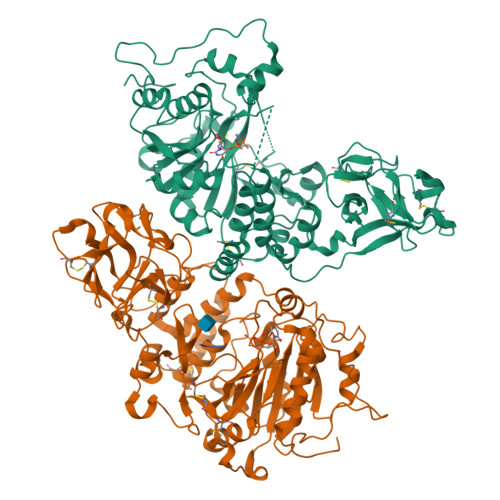
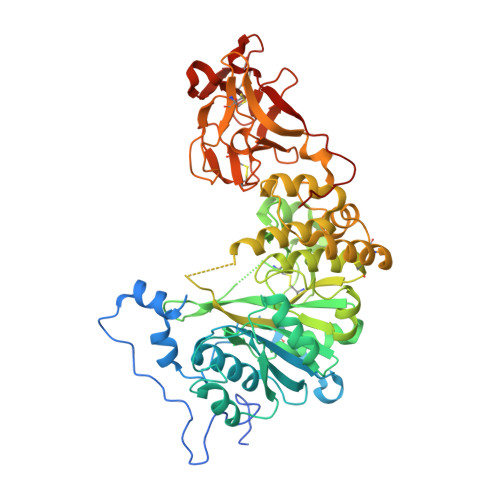
A Toxoplasma gondii O-glycosyltransferase that modulates bradyzoite cyst wall rigidity is distinct from host homologues.
Kumar, P., Tomita, T., Gerken, T.A., Ballard, C.J., Lee, Y.S., Weiss, L.M., Samara, N.L.(2024) Nat Commun 15: 3792-3792
- PubMed: 38710711
- DOI: https://doi.org/10.1038/s41467-024-48253-w
- Primary Citation of Related Structures:
8UHV, 8UHZ, 8UI1, 8UI6, 8UJE, 8UJF, 8UJG, 8UJH - PubMed Abstract:
Infection with the apicomplexan protozoan Toxoplasma gondii can be life-threatening in immunocompromised hosts. Transmission frequently occurs through the oral ingestion of T. gondii bradyzoite cysts, which transition to tachyzoites, disseminate, and then form cysts containing bradyzoites in the central nervous system, resulting in latent infection. Encapsulation of bradyzoites by a cyst wall is critical for immune evasion, survival, and transmission. O-glycosylation of the protein CST1 by the mucin-type O-glycosyltransferase T. gondii (Txg) GalNAc-T3 influences cyst wall rigidity and stability. Here, we report X-ray crystal structures of TxgGalNAc-T3, revealing multiple features that are strictly conserved among its apicomplexan homologues. This includes a unique 2 nd metal that is coupled to substrate binding and enzymatic activity in vitro and cyst wall O-glycosylation in T. gondii. The study illustrates the divergence of pathogenic protozoan GalNAc-Ts from their host homologues and lays the groundwork for studying apicomplexan GalNAc-Ts as therapeutic targets in disease.
Organizational Affiliation:
Structural Biochemistry Unit, National Institute of Dental and Craniofacial Research, NIH, Bethesda, MD, 20892, USA.