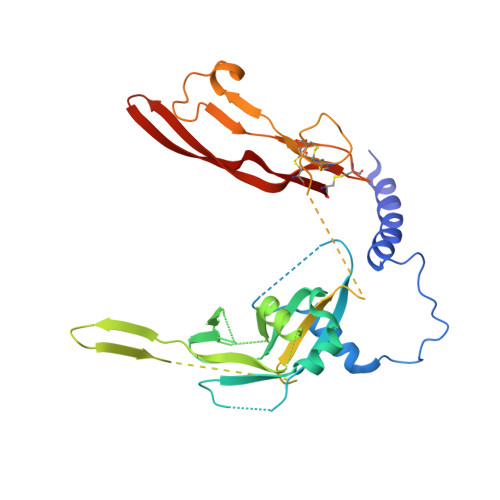
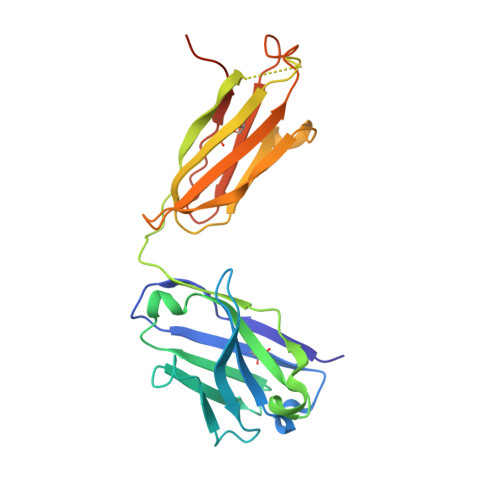
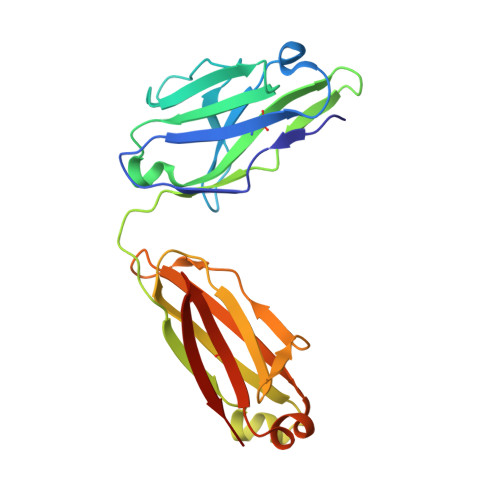
An antibody that inhibits TGF-beta 1 release from latent extracellular matrix complexes attenuates the progression of renal fibrosis.
Jackson, J.W., Streich Jr., F.C., Pal, A., Coricor, G., Boston, C., Brueckner, C.T., Canonico, K., Chapron, C., Cote, S., Dagbay, K.B., Danehy Jr., F.T., Kavosi, M., Kumar, S., Lin, S., Littlefield, C., Looby, K., Manohar, R., Martin, C.J., Wood, M., Zawadzka, A., Wawersik, S., Nicholls, S.B., Datta, A., Buckler, A., Schurpf, T., Carven, G.J., Qatanani, M., Fogel, A.I.(2024) Sci Signal 17: eadn6052-eadn6052
- PubMed: 38980922
- DOI: https://doi.org/10.1126/scisignal.adn6052
- Primary Citation of Related Structures:
8UDZ - PubMed Abstract:
Inhibitors of the transforming growth factor-β (TGF-β) pathway are potentially promising antifibrotic therapies, but nonselective simultaneous inhibition of all three TGF-β homologs has safety liabilities. TGF-β1 is noncovalently bound to a latency-associated peptide that is, in turn, covalently bound to different presenting molecules within large latent complexes. The latent TGF-β-binding proteins (LTBPs) present TGF-β1 in the extracellular matrix, and TGF-β1 is presented on immune cells by two transmembrane proteins, glycoprotein A repetitions predominant (GARP) and leucine-rich repeat protein 33 (LRRC33). Here, we describe LTBP-49247, an antibody that selectively bound to and inhibited the activation of TGF-β1 presented by LTBPs but did not bind to TGF-β1 presented by GARP or LRRC33. Structural studies demonstrated that LTBP-49247 recognized an epitope on LTBP-presented TGF-β1 that is not accessible on GARP- or LRRC33-presented TGF-β1, explaining the antibody's selectivity for LTBP-complexed TGF-β1. In two rodent models of kidney fibrosis of different etiologies, LTBP-49247 attenuated fibrotic progression, indicating the central role of LTBP-presented TGF-β1 in renal fibrosis. In mice, LTBP-49247 did not have the toxic effects associated with less selective TGF-β inhibitors. These results establish the feasibility of selectively targeting LTBP-bound TGF-β1 as an approach for treating fibrosis.
Organizational Affiliation:
Scholar Rock Inc., 301 Binney Street, Cambridge, MA 02142, USA.