High-resolution double vision of the allosteric phosphatase PTP1B.
Sharma, S., Skaist Mehlman, T., Sagabala, R.S., Boivin, B., Keedy, D.A.(2024) Acta Crystallogr F Struct Biol Commun 80: 1-12
- PubMed: 38133579
- DOI: https://doi.org/10.1107/S2053230X23010749
- Primary Citation of Related Structures:
8U1E - PubMed Abstract:
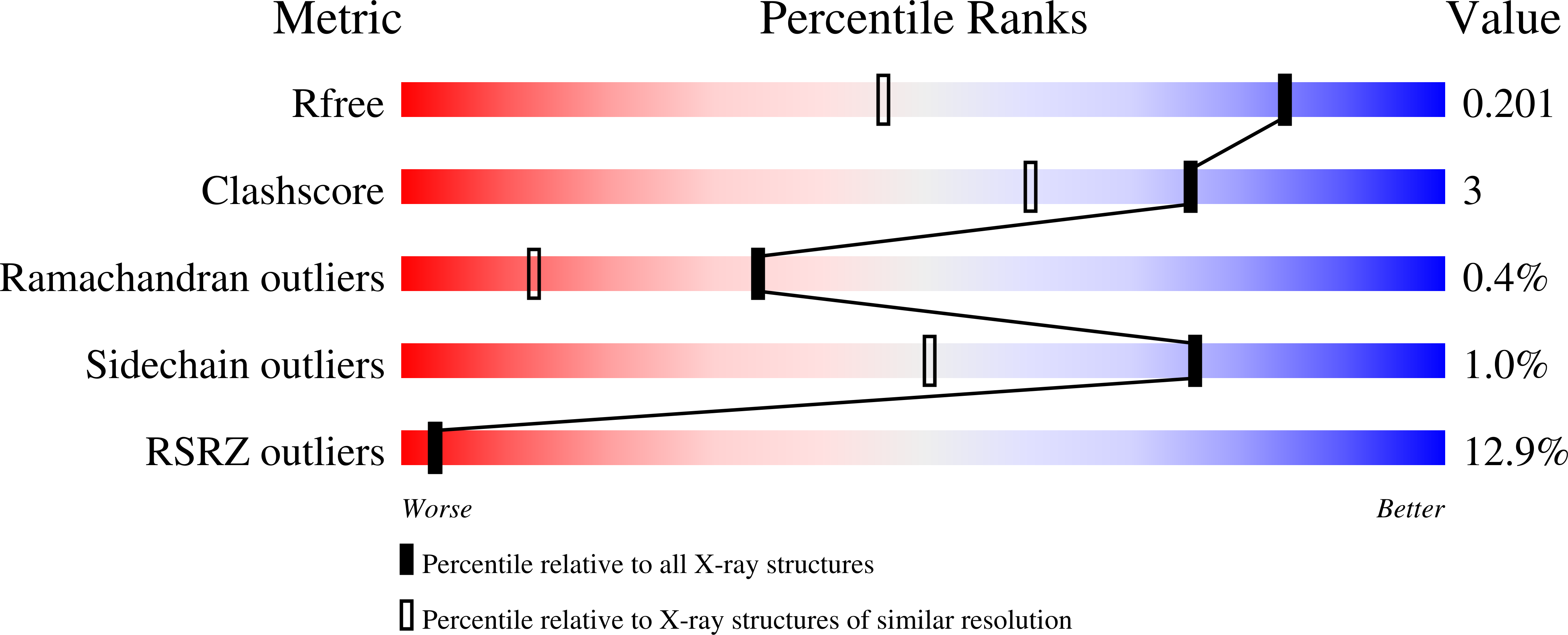
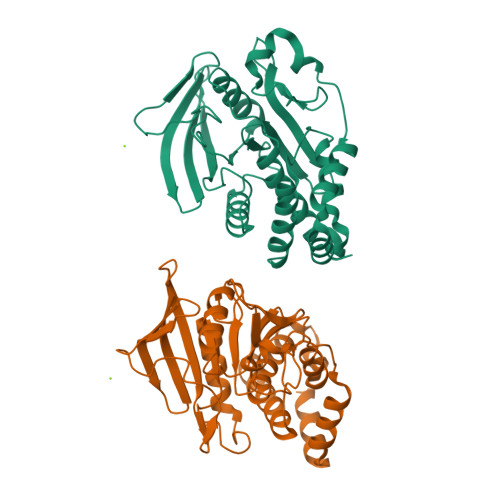
Protein tyrosine phosphatase 1B (PTP1B) plays important roles in cellular homeostasis and is a highly validated therapeutic target for multiple human ailments, including diabetes, obesity and breast cancer. However, much remains to be learned about how conformational changes may convey information through the structure of PTP1B to enable allosteric regulation by ligands or functional responses to mutations. High-resolution X-ray crystallography can offer unique windows into protein conformational ensembles, but comparison of even high-resolution structures is often complicated by differences between data sets, including non-isomorphism. Here, the highest resolution crystal structure of apo wild-type (WT) PTP1B to date is presented out of a total of ∼350 PTP1B structures in the PDB. This structure is in a crystal form that is rare for PTP1B, with two unique copies of the protein that exhibit distinct patterns of conformational heterogeneity, allowing a controlled comparison of local disorder across the two chains within the same asymmetric unit. The conformational differences between these chains are interrogated in the apo structure and between several recently reported high-resolution ligand-bound structures. Electron-density maps in a high-resolution structure of a recently reported activating double mutant are also examined, and unmodeled alternate conformations in the mutant structure are discovered that coincide with regions of enhanced conformational heterogeneity in the new WT structure. These results validate the notion that these mutations operate by enhancing local dynamics, and suggest a latent susceptibility to such changes in the WT enzyme. Together, these new data and analysis provide a detailed view of the conformational ensemble of PTP1B and highlight the utility of high-resolution crystallography for elucidating conformational heterogeneity with potential relevance for function.
Organizational Affiliation:
Structural Biology Initiative, CUNY Advanced Science Research Center, New York, NY 10031, USA.