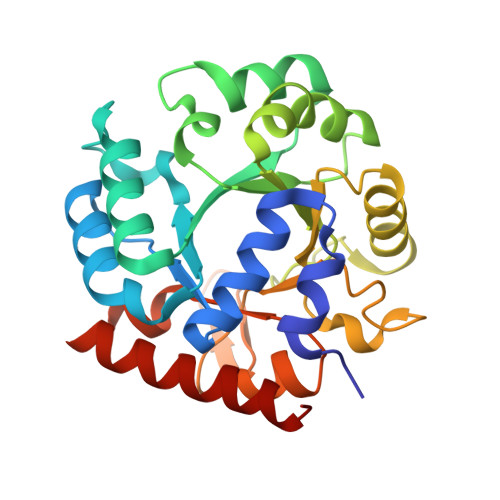
Structural and functional properties of uridine 5'-monophosphate synthase from Coffea arabica.
Hinojosa-Cruz, A., Diaz-Sanchez, A.G., Diaz-Vilchis, A., Gonzalez-Segura, L.(2024) Int J Biol Macromol 259: 129226-129226
- PubMed: 38184030
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.129226
- Primary Citation of Related Structures:
8U0Z - PubMed Abstract:
In higher eukaryotes and plants, the last two sequential steps in the de novo biosynthesis of uridine 5'-monophosphate (UMP) are catalyzed by a bifunctional natural chimeric protein called UMP synthase (UMPS). In higher plants, UMPS consists of two naturally fused enzymes: orotate phosphoribosyltransferase (OPRTase) at N-terminal and orotidine-5'-monophosphate decarboxylase (ODCase) at C-terminal. In this work, we obtained the full functional recombinant protein UMPS from Coffea arabica (CaUMPS) and studied its structure-function relationships. A biochemical and structural characterization of a plant UMPS with its two functional domains is described together with the presentation of the first crystal structure of a plant ODCase at 1.4 Å resolution. The kinetic parameters measured of CaOPRTase and CaODCase domains were comparable to those reported. The crystallographic structure revealed that CaODCase is a dimer that conserves the typical fold observed in other ODCases from prokaryote and eukaryote with a 1-deoxy-ribofuranose-5'-phosphate molecule bound in the active site of one subunit induced a closed conformation. Our results add to the knowledge of one of the key enzymes of the de novo biosynthesis of pyrimidines in plant metabolism and open the door to future applications.
Organizational Affiliation:
Departamento de Bioquímica, Facultad de Química, Universidad Nacional Autónoma de México, Ciudad de México 04510, Mexico.