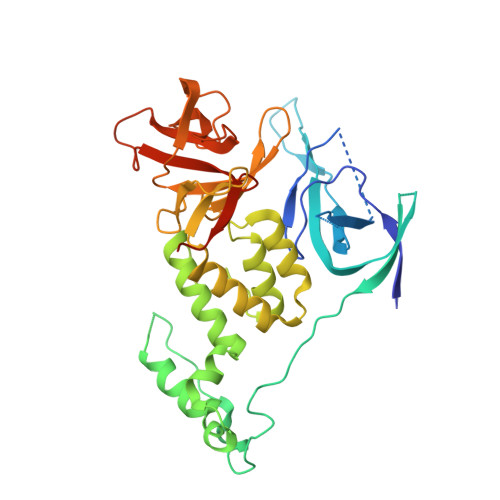
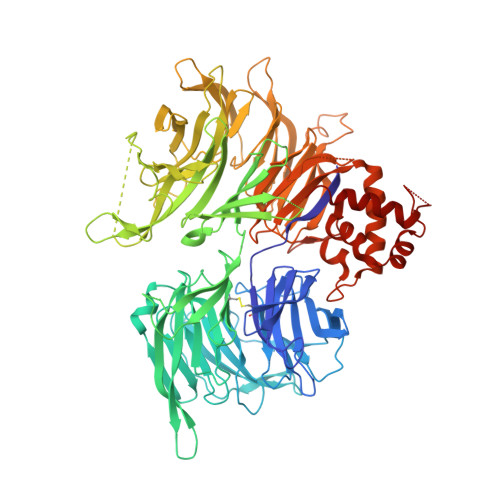
A molecular glue degrader of the WIZ transcription factor for fetal hemoglobin induction.
Ting, P.Y., Borikar, S., Kerrigan, J.R., Thomsen, N.M., Aghania, E., Hinman, A.E., Reyes, A., Pizzato, N., Fodor, B.D., Wu, F., Belew, M.S., Mao, X., Wang, J., Chitnis, S., Niu, W., Hachey, A., Cobb, J.S., Savage, N.A., Burke, A., Paulk, J., Dovala, D., Lin, J., Clifton, M.C., Ornelas, E., Ma, X., Ware, N.F., Sanchez, C.C., Taraszka, J., Terranova, R., Knehr, J., Altorfer, M., Barnes, S.W., Beckwith, R.E.J., Solomon, J.M., Dales, N.A., Patterson, A.W., Wagner, J., Bouwmeester, T., Dranoff, G., Stevenson, S.C., Bradner, J.E.(2024) Science 385: 91-99
- PubMed: 38963839
- DOI: https://doi.org/10.1126/science.adk6129
- Primary Citation of Related Structures:
8TZX - PubMed Abstract:
Sickle cell disease (SCD) is a prevalent, life-threatening condition attributable to a heritable mutation in β-hemoglobin. Therapeutic induction of fetal hemoglobin (HbF) can ameliorate disease complications and has been intently pursued. However, safe and effective small-molecule inducers of HbF remain elusive. We report the discovery of dWIZ-1 and dWIZ-2, molecular glue degraders of the WIZ transcription factor that robustly induce HbF in erythroblasts. Phenotypic screening of a cereblon (CRBN)-biased chemical library revealed WIZ as a previously unknown repressor of HbF. WIZ degradation is mediated by recruitment of WIZ(ZF7) to CRBN by dWIZ-1, as resolved by crystallography of the ternary complex. Pharmacological degradation of WIZ was well tolerated and induced HbF in humanized mice and cynomolgus monkeys. These findings establish WIZ degradation as a globally accessible therapeutic strategy for SCD.
Organizational Affiliation:
Novartis Biomedical Research, Cambridge, MA, USA.