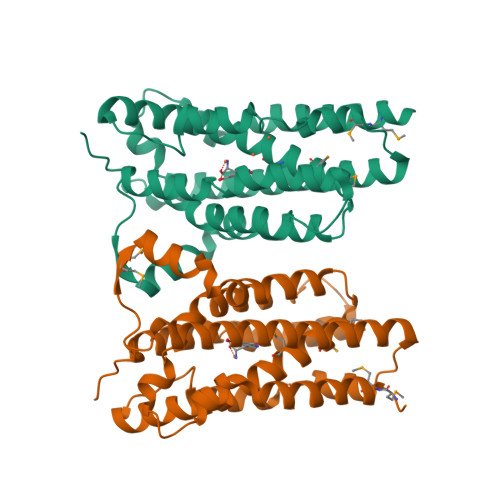
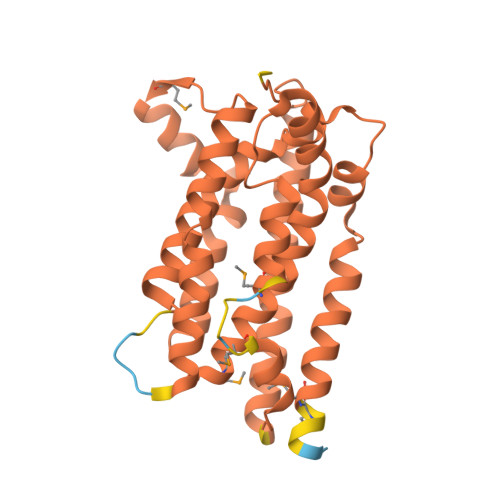
A single diiron enzyme catalyses the oxidative rearrangement of tryptophan to indole nitrile.
Adak, S., Ye, N., Calderone, L.A., Duan, M., Lubeck, W., Schafer, R.J.B., Lukowski, A.L., Houk, K.N., Pandelia, M.E., Drennan, C.L., Moore, B.S.(2024) Nat Chem 16: 1989-1998
- PubMed: 39285206
- DOI: https://doi.org/10.1038/s41557-024-01603-z
- Primary Citation of Related Structures:
8TWN, 8TWT, 8TWW - PubMed Abstract:
Nitriles are uncommon in nature and are typically constructed from oximes through the oxidative decarboxylation of amino acid substrates or from the derivatization of carboxylic acids. Here we report a third nitrile biosynthesis strategy featuring the cyanobacterial nitrile synthase AetD. During the biosynthesis of the eagle-killing neurotoxin, aetokthonotoxin, AetD transforms the 2-aminopropionate portion of 5,7-dibromo-L-tryptophan to a nitrile. Employing a combination of structural, biochemical and biophysical techniques, we characterized AetD as a non-haem diiron enzyme that belongs to the emerging haem-oxygenase-like dimetal oxidase superfamily. High-resolution crystal structures of AetD together with the identification of catalytically relevant products provide mechanistic insights into how AetD affords this unique transformation, which we propose proceeds via an aziridine intermediate. Our work presents a unique template for nitrile biogenesis and portrays a substrate binding and metallocofactor assembly mechanism that may be shared among other haem-oxygenase-like dimetal oxidase enzymes.
Organizational Affiliation:
Center for Marine Biotechnology and Biomedicine, Scripps Institution of Oceanography, University of California San Diego, La Jolla, CA, USA.