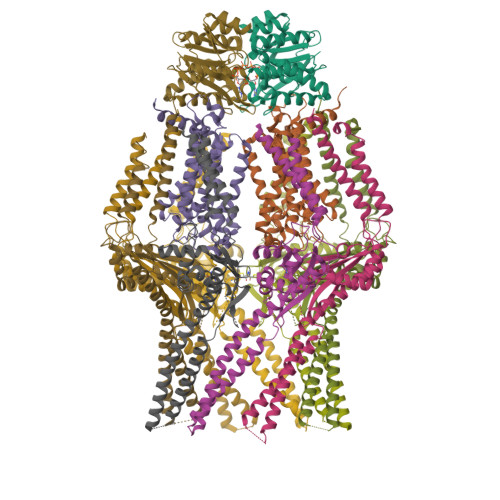
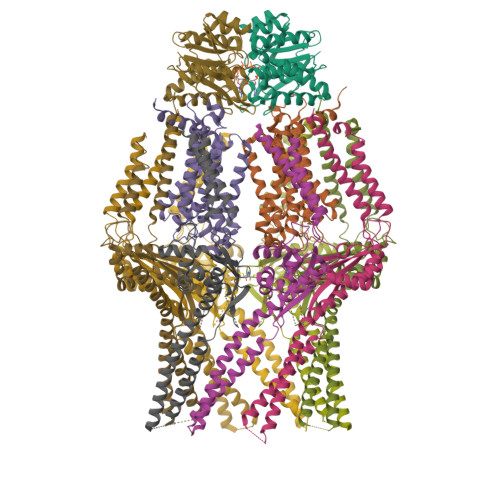
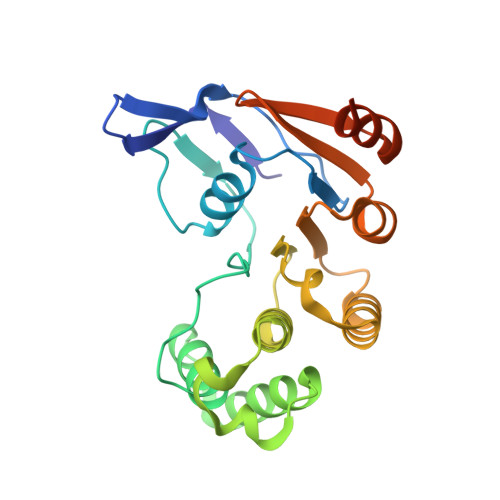
Molecular insights into capsular polysaccharide secretion.
Kuklewicz, J., Zimmer, J.(2024) Nature 628: 901-909
- PubMed: 38570679
- DOI: https://doi.org/10.1038/s41586-024-07248-9
- Primary Citation of Related Structures:
8TSH, 8TSI, 8TSL, 8TSW, 8TT3, 8TUN - PubMed Abstract:
Capsular polysaccharides (CPSs) fortify the cell boundaries of many commensal and pathogenic bacteria 1 . Through the ABC-transporter-dependent biosynthesis pathway, CPSs are synthesized intracellularly on a lipid anchor and secreted across the cell envelope by the KpsMT ABC transporter associated with the KpsE and KpsD subunits 1,2 . Here we use structural and functional studies to uncover crucial steps of CPS secretion in Gram-negative bacteria. We show that KpsMT has broad substrate specificity and is sufficient for the translocation of CPSs across the inner bacterial membrane, and we determine the cell surface organization and localization of CPSs using super-resolution fluorescence microscopy. Cryo-electron microscopy analyses of the KpsMT-KpsE complex in six different states reveal a KpsE-encaged ABC transporter, rigid-body conformational rearrangements of KpsMT during ATP hydrolysis and recognition of a glycolipid inside a membrane-exposed electropositive canyon. In vivo CPS secretion assays underscore the functional importance of canyon-lining basic residues. Combined, our analyses suggest a molecular model of CPS secretion by ABC transporters.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia School of Medicine, Charlottesville, VA, USA.