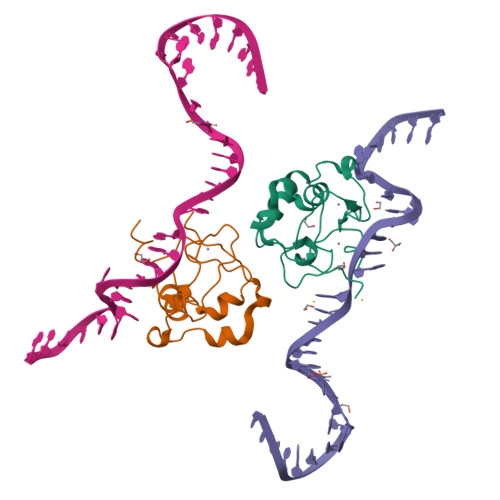
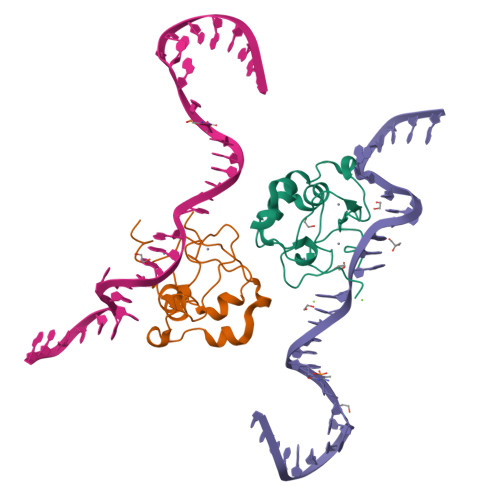
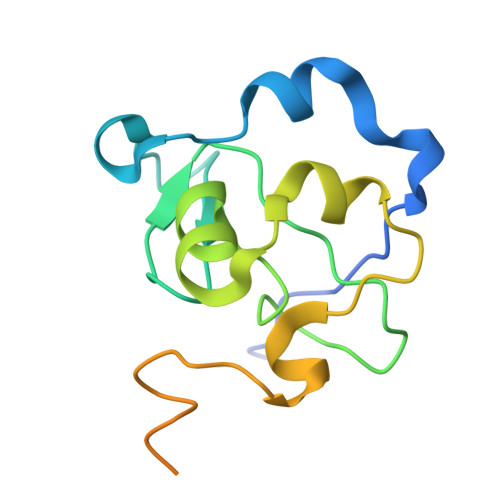
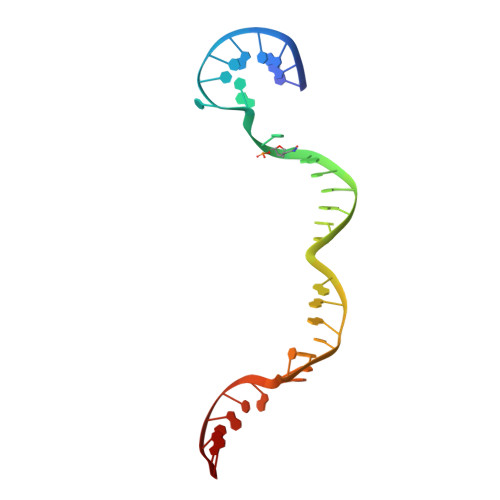
The ICF syndrome protein CDCA7 harbors a unique DNA-binding domain that recognizes a CpG dyad in the context of a non-B DNA.
Hardikar, S., Ren, R., Ying, Z., Horton, J.R., Bramble, M.D., Liu, B., Lu, Y., Liu, B., Dan, J., Zhang, X., Cheng, X., Chen, T.(2023) bioRxiv
- PubMed: 38168392
- DOI: https://doi.org/10.1101/2023.12.15.571946
- Primary Citation of Related Structures:
8TLE, 8TLF, 8TLG, 8TLH, 8TLJ, 8TLK - PubMed Abstract:
CDCA7 , encoding a protein with a C-terminal cysteine-rich domain (CRD), is mutated in immunodeficiency, centromeric instability and facial anomalies (ICF) syndrome, a disease related to hypomethylation of juxtacentromeric satellite DNA. How CDCA7 directs DNA methylation to juxtacentromeric regions is unknown. Here, we show that the CDCA7 CRD adopts a unique zinc-binding structure that recognizes a CpG dyad in a non-B DNA formed by two sequence motifs. CDCA7, but not ICF mutants, preferentially binds the non-B DNA with strand-specific CpG hemi-methylation. The unmethylated sequence motif is highly enriched at centromeres of human chromosomes, whereas the methylated motif is distributed throughout the genome. At S phase, CDCA7, but not ICF mutants, is concentrated in constitutive heterochromatin foci, and the formation of such foci can be inhibited by exogenous hemi-methylated non-B DNA bound by the CRD. Binding of the non-B DNA formed in juxtacentromeric regions during DNA replication provides a mechanism by which CDCA7 controls the specificity of DNA methylation.