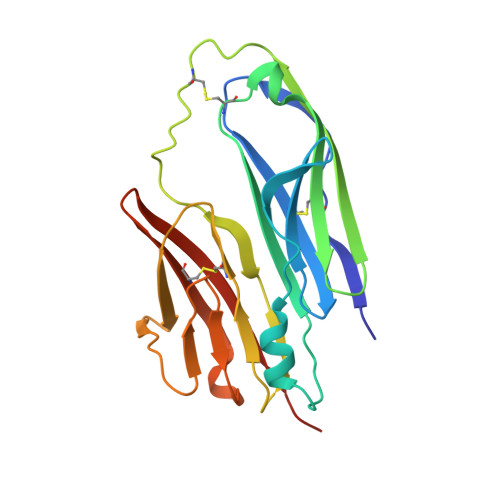
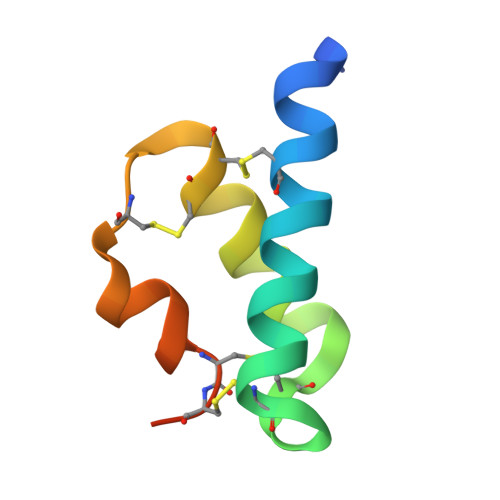
Nematode Extracellular Protein Interactome Expands Connections between Signaling Pathways.
Nawrocka, W.I., Cheng, S., Hao, B., Rosen, M.C., Cortes, E., Baltrusaitis, E.E., Aziz, Z., Kovacs, I.A., Ozkan, E.(2024) bioRxiv
- PubMed: 39026773
- DOI: https://doi.org/10.1101/2024.07.08.602367
- Primary Citation of Related Structures:
8TK9, 8TKT, 8TKU - PubMed Abstract:
Multicellularity was accompanied by the emergence of new classes of cell surface and secreted proteins. The nematode C. elegans is a favorable model to study cell surface interactomes, given its well-defined and stereotyped cell types and intercellular contacts. Here we report our C. elegans extracellular interactome dataset, the largest yet for an invertebrate. Most of these interactions were unknown, despite recent datasets for flies and humans, as our collection contains a larger selection of protein families. We uncover new interactions for all four major axon guidance pathways, including ectodomain interactions between three of the pathways. We demonstrate that a protein family known to maintain axon locations are secreted receptors for insulins. We reveal novel interactions of cystine-knot proteins with putative signaling receptors, which may extend the study of neurotrophins and growth-factor-mediated functions to nematodes. Finally, our dataset provides insights into human disease mechanisms and how extracellular interactions may help establish connectomes.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA.