CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO GPZ6-a.01 FAB
Morano, N.C., Becker, J.E., Shapiro, L.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Envelope glycoprotein gp160 | A [auth G], C [auth N], E [auth O] | 480 | Human immunodeficiency virus 1 | Mutation(s): 0 Gene Names: env | 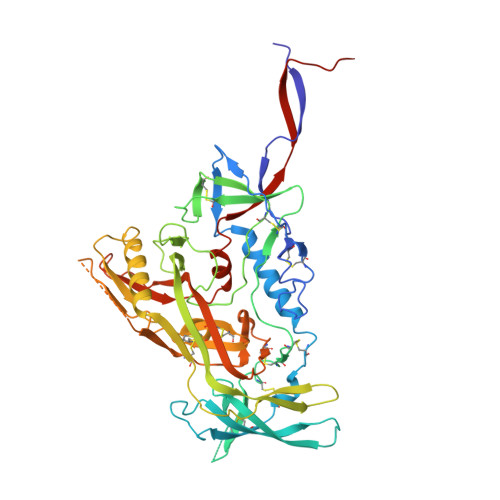 |
UniProt | |||||
Find proteins for Q2N0S6 (Human immunodeficiency virus 1) Explore Q2N0S6 Go to UniProtKB: Q2N0S6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2N0S6 | ||||
Glycosylation | |||||
| Glycosylation Sites: 13 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Envelope glycoprotein gp41 | B, D [auth H], F [auth I] | 153 | Human immunodeficiency virus 1 | Mutation(s): 2 Gene Names: env | 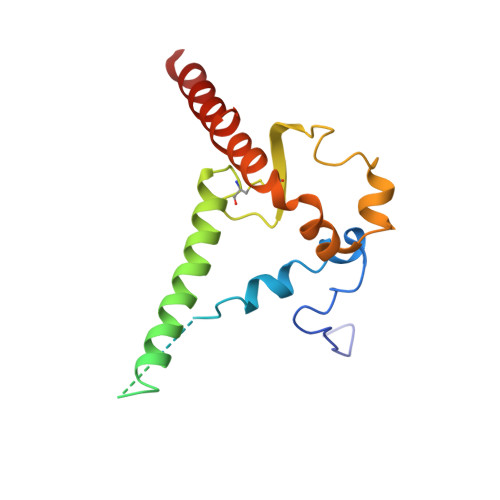 |
UniProt | |||||
Find proteins for Q2N0S6 (Human immunodeficiency virus 1) Explore Q2N0S6 Go to UniProtKB: Q2N0S6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2N0S6 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antibody GPZ6-a.01 Heavy Chain | G [auth Q], I [auth P], K [auth R] | 126 | Macaca | Mutation(s): 0 | 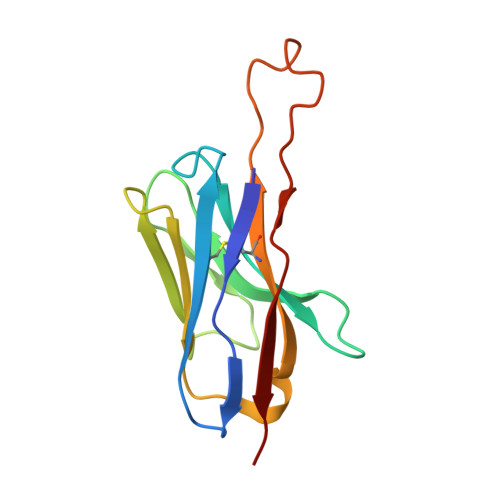 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antibody GPZ6-a.01 Light Chain | H [auth T], J [auth S], L [auth U] | 107 | Macaca | Mutation(s): 0 | 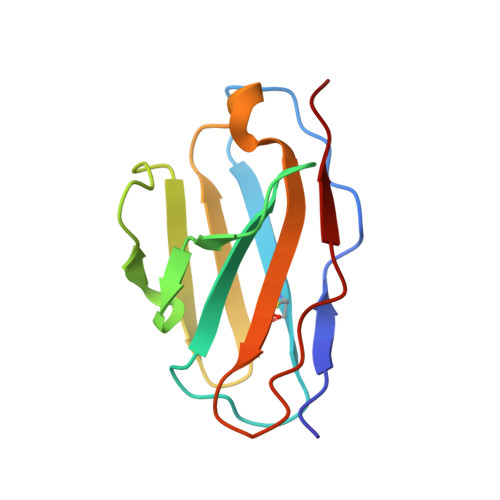 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | M [auth A], P [auth E], S [auth F] | 6 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G56014GC GlyCosmos: G56014GC GlyGen: G56014GC | |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | N [auth C], O [auth D], Q [auth J], R [auth L], T [auth K], N [auth C], O [auth D], Q [auth J], R [auth L], T [auth K], U [auth M] | 2 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | AA [auth G] AB [auth H] BA [auth G] BB [auth O] CA [auth G] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | -- |