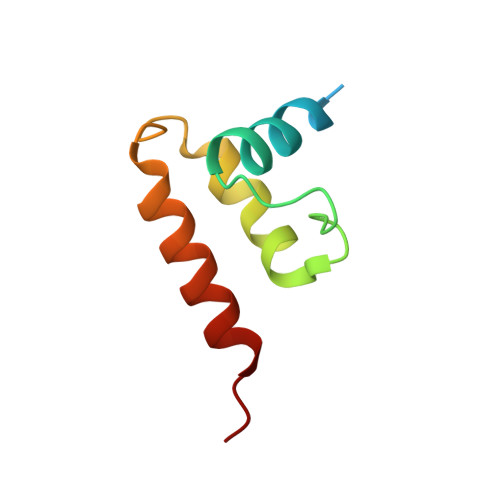
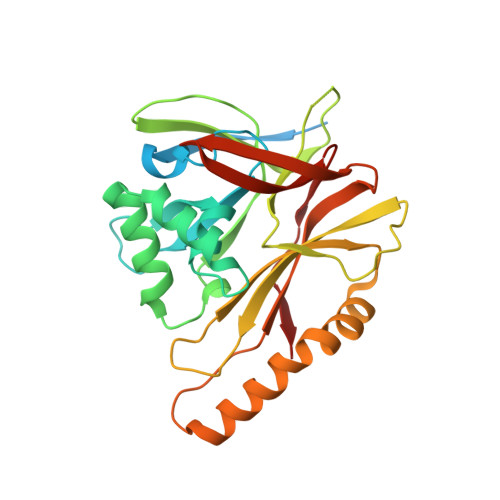
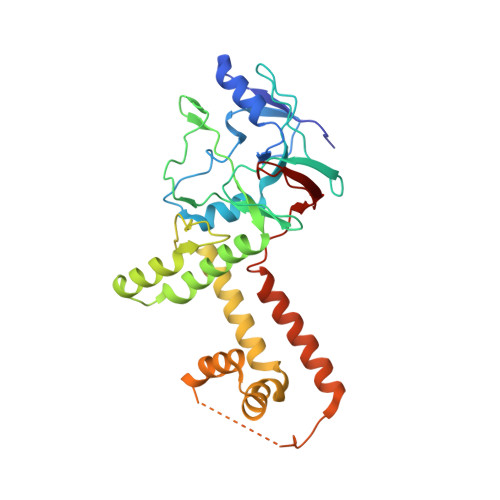
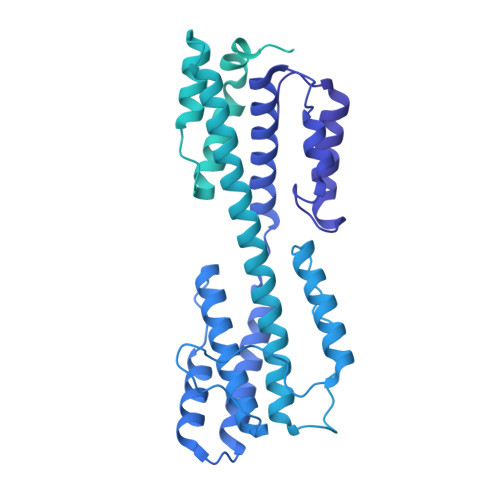
The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM.
Jih, J., Liu, Y.T., Liu, W., Zhou, Z.H.(2024) Sci Adv 10: eadj1640-eadj1640
- PubMed: 38394211
- DOI: https://doi.org/10.1126/sciadv.adj1640
- Primary Citation of Related Structures:
8TEP, 8TES, 8TET, 8TEU, 8TEW - PubMed Abstract:
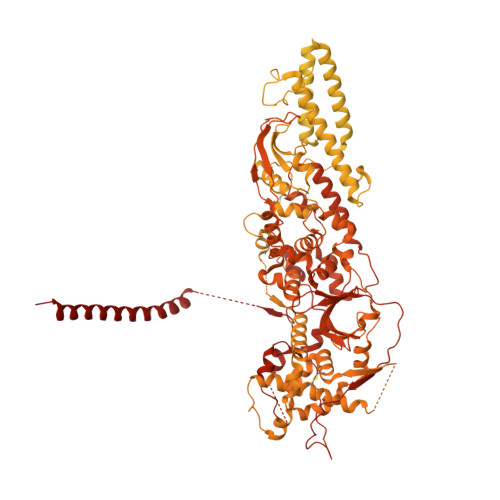
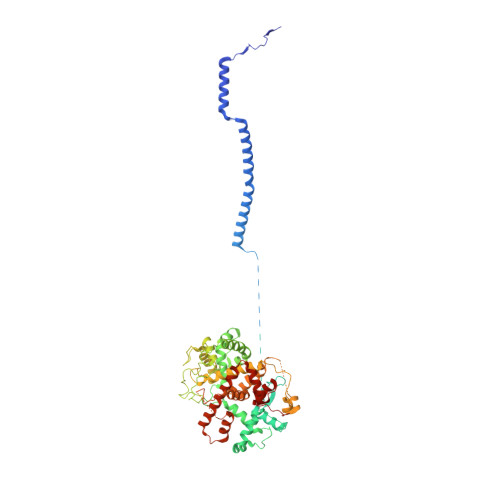
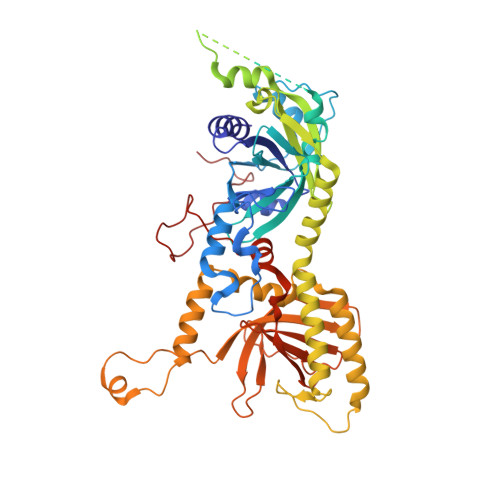
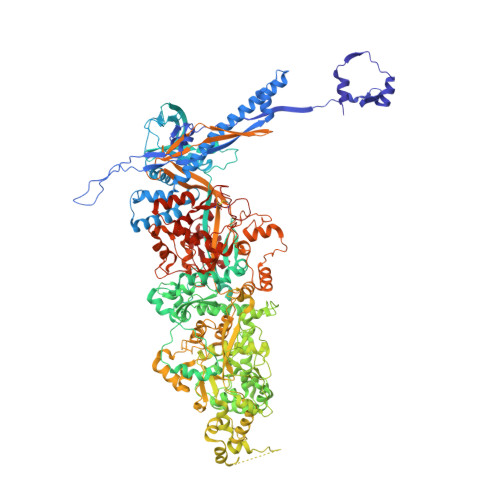
The compartmentalization of eukaryotic cells presents considerable challenges to the herpesvirus life cycle. The herpesvirus tegument, a bulky proteinaceous aggregate sandwiched between herpesviruses' capsid and envelope, is uniquely evolved to address these challenges, yet tegument structure and organization remain poorly characterized. We use deep-learning-enhanced cryogenic electron microscopy to investigate the tegument of human cytomegalovirus virions and noninfectious enveloped particles (NIEPs; a genome packaging-aborted state), revealing a portal-biased tegumentation scheme. We resolve atomic structures of portal vertex-associated tegument (PVAT) and identify multiple configurations of PVAT arising from layered reorganization of pUL77, pUL48 (large tegument protein), and pUL47 (inner tegument protein) assemblies. Analyses show that pUL77 seals the last-packaged viral genome end through electrostatic interactions, pUL77 and pUL48 harbor a head-linker-capsid-binding motif conducive to PVAT reconfiguration, and pUL47/48 dimers form 45-nm-long filaments extending from the portal vertex. These results provide a structural framework for understanding how herpesvirus tegument facilitates and evolves during processes spanning viral genome packaging to delivery.
Organizational Affiliation:
Molecular Biology Institute, University of California, Los Angeles (UCLA), Los Angeles, CA 90095, USA.