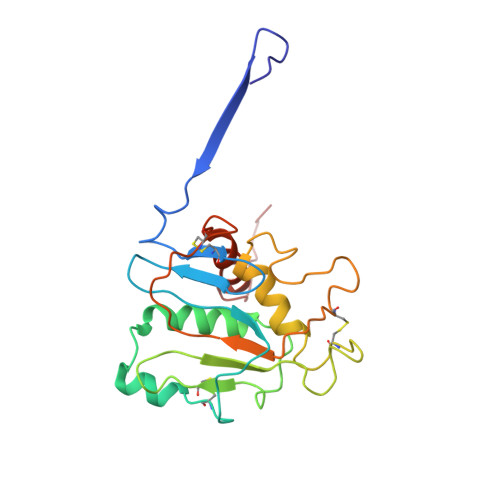
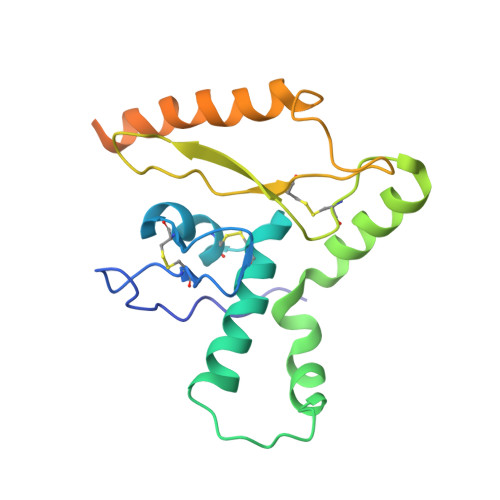
Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Gorman, J., Cheung, C.S., Duan, Z., Ou, L., Wang, M., Chen, X., Cheng, C., Biju, A., Sun, Y., Wang, P., Yang, Y., Zhang, B., Boyington, J.C., Bylund, T., Charaf, S., Chen, S.J., Du, H., Henry, A.R., Liu, T., Sarfo, E.K., Schramm, C.A., Shen, C.H., Stephens, T., Teng, I.T., Todd, J.P., Tsybovsky, Y., Verardi, R., Wang, D., Wang, S., Wang, Z., Zheng, C.Y., Zhou, T., Douek, D.C., Mascola, J.R., Ho, D.D., Ho, M., Kwong, P.D.(2024) Nat Commun 15: 285-285
- PubMed: 38177144
- DOI: https://doi.org/10.1038/s41467-023-44534-y
- Primary Citation of Related Structures:
8T5C - PubMed Abstract:
Lassa virus (LASV) infection is expanding outside its traditionally endemic areas in West Africa, posing a pandemic biothreat. LASV-neutralizing antibodies, moreover, have proven difficult to elicit. To gain insight into LASV neutralization, here we develop a prefusion-stabilized LASV glycoprotein trimer (GPC), pan it against phage libraries comprising single-domain antibodies (nanobodies) from shark and camel, and identify one, D5, which neutralizes LASV. Cryo-EM analyses reveal D5 to recognize a cleavage-dependent site-of-vulnerability at the trimer apex. The recognized site appears specific to GPC intermediates, with protomers lacking full cleavage between GP1 and GP2 subunits. Guinea pig immunizations with the prefusion-stabilized cleavage-intermediate LASV GPC, first as trimer and then as a nanoparticle, induce neutralizing responses, targeting multiple epitopes including that of D5; we identify a neutralizing antibody (GP23) from the immunized guinea pigs. Collectively, our findings define a prefusion-stabilized GPC trimer, reveal an apex-situated site-of-vulnerability, and demonstrate elicitation of LASV-neutralizing responses by a cleavage-intermediate LASV trimer.
Organizational Affiliation:
Vaccine Research Center, National Institutes of Health, Bethesda, MD, 20892, USA.