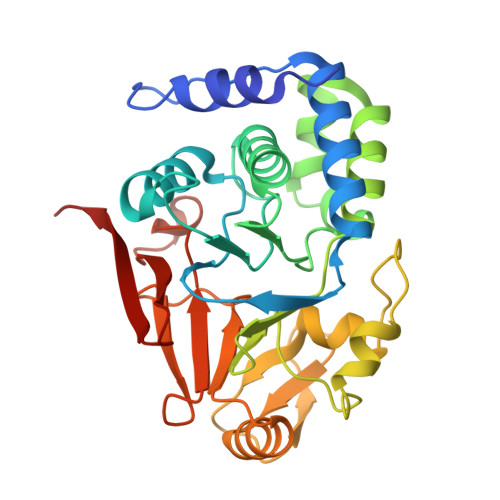
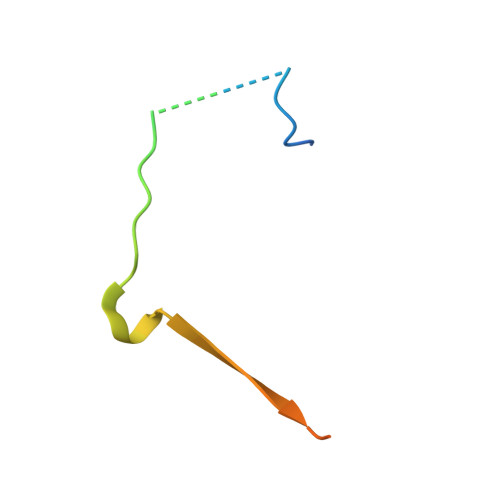
A protein phosphatase 1 specific phos phatase ta rgeting p eptide (PhosTAP) to identify the PP1 phosphatome.
Choy, M.S., Nguyen, H.T., Kumar, G.S., Peti, W., Kettenbach, A.N., Page, R.(2024) Proc Natl Acad Sci U S A 121: e2415383121-e2415383121
- PubMed: 39446389
- DOI: https://doi.org/10.1073/pnas.2415383121
- Primary Citation of Related Structures:
8SW5, 8SW6 - PubMed Abstract:
Phosphoprotein phosphatases (PPPs) are the key serine/threonine phosphatases that regulate all essential signaling cascades. In particular, Protein Phosphatase 1 (PP1) dephosphorylates ~80% of all ser/thr phosphorylation sites. Here, we developed a phosphatase targeting peptide (PhosTAP) that binds all PP1 isoforms and does so with a stronger affinity than any other known PP1 regulator. This PhosTAP can be used as a PP1 recruitment tool for Phosphorylation Targeting Chimera (PhosTAC)-type recruitment in in vitro and cellular experiments, as well as in phosphoproteomics experiments to identify PP1-specific substrates and phosphosites. The latter is especially important to further our understanding of cellular signaling, as the identification of substrates and especially phosphosites that are targeted by specific phosphatases lags behind that of their kinase counterparts. Using PhosTAP-based proteomics, we show that, counter to our current understanding, many PP1 regulators are also substrates, that the number of residues between regulator PP1-binding and phosphosites vary significantly, and that PP1 counteracts the activities of mitotic kinases. Finally, we also found that Haspin kinase is a direct substrate of PP1 and that its PP1-dependent dephosphorylation modulates its activity during anaphase. Together, we show that PP1-specific PhosTAPs are a powerful tool for +studying PP1 activity in vitro and in cells.
Organizational Affiliation:
Department of Molecular Biology and Biophysics, UConn Health, Farmington, CT 06030.