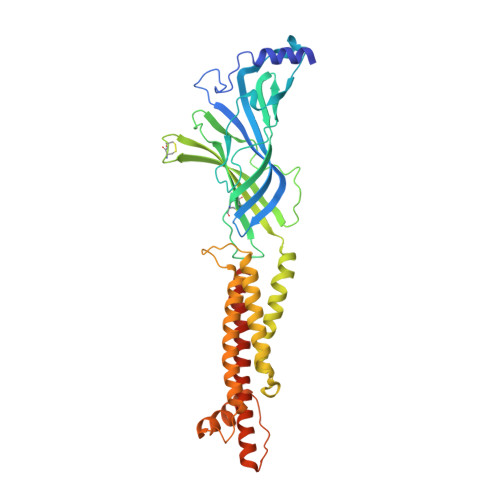
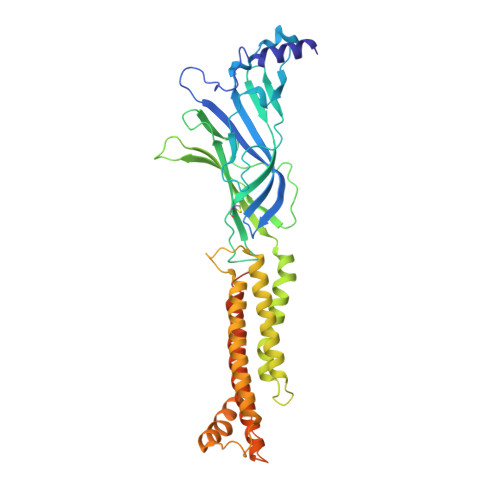
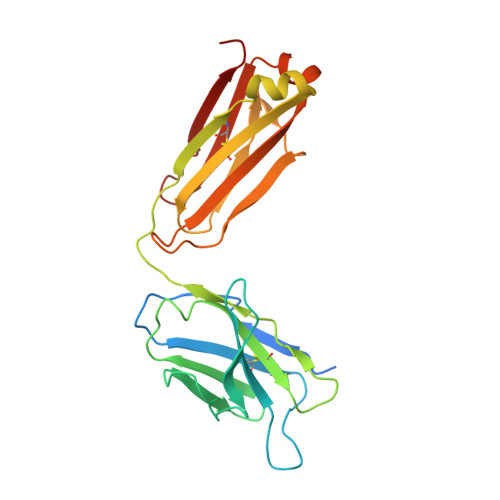
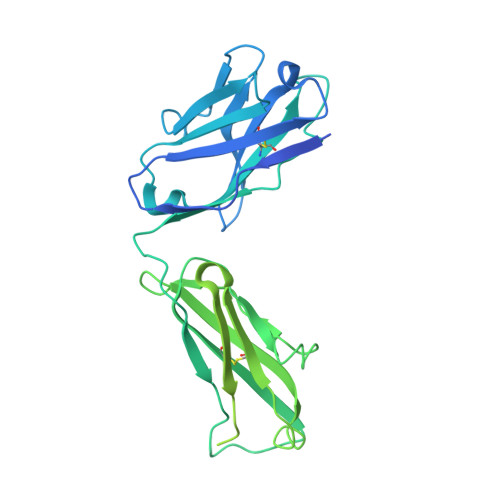
Structural bases for stoichiometry-selective calcium potentiation of a neuronal nicotinic receptor.
Mazzaferro, S., Kang, G., Natarajan, K., Hibbs, R.E., Sine, S.M.(2024) Br J Pharmacol 181: 1973-1992
- PubMed: 38454578
- DOI: https://doi.org/10.1111/bph.16321
- Primary Citation of Related Structures:
8SSZ, 8ST0, 8ST1, 8ST2, 8ST3, 8ST4 - PubMed Abstract:
α4β2 nicotinic acetylcholine (nACh) receptors assemble in two stoichiometric forms, one of which is potentiated by calcium. The sites of calcium binding that underpin potentiation are not known. To identify calcium binding sites, we applied cryo-electron microscopy (cryo-EM) and molecular dynamics (MD) simulations to each stoichiometric form of the α4β2 nACh receptor in the presence of calcium ions. To test whether the identified calcium sites are linked to potentiation, we generated mutants of anionic residues at the sites, expressed wild type and mutant receptors in clonal mammalian fibroblasts, and recorded ACh-elicited single-channel currents with or without calcium. Both cryo-EM and MD simulations show calcium bound to a site between the extracellular and transmembrane domains of each α4 subunit (ECD-TMD site). Substituting alanine for anionic residues at the ECD-TMD site abolishes stoichiometry-selective calcium potentiation, as monitored by single-channel patch clamp electrophysiology. Additionally, MD simulation reveals calcium association at subunit interfaces within the extracellular domain. Substituting alanine for anionic residues at the ECD sites reduces or abolishes stoichiometry-selective calcium potentiation. Stoichiometry-selective calcium potentiation of the α4β2 nACh receptor is achieved by calcium association with topographically distinct sites framed by anionic residues within the α4 subunit and between the α4 and β2 subunits. Stoichiometry-selective calcium potentiation could result from the greater number of calcium sites in the stoichiometric form with three rather than two α4 subunits. The results are relevant to modulation of signalling via α4β2 nACh receptors in physiological and pathophysiological conditions.
Organizational Affiliation:
Department of Physiology and Biomedical Engineering, Mayo Clinic, Rochester, Minnesota, USA.