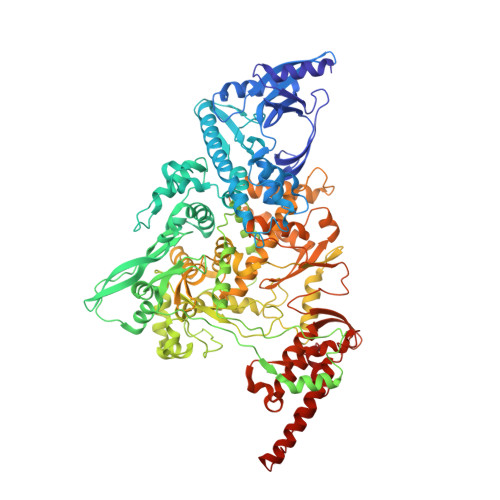
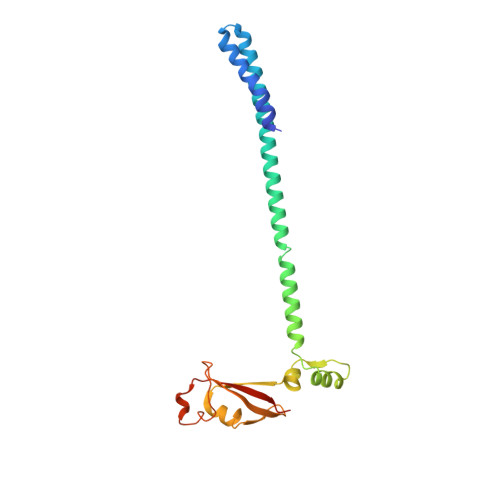
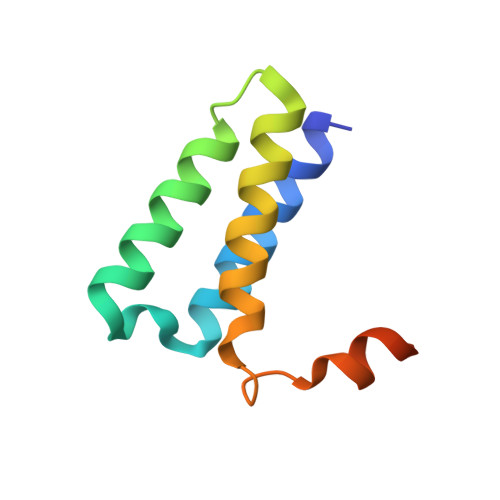
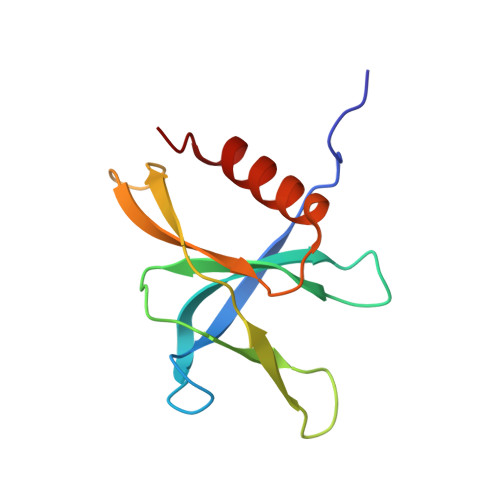
Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Small, G.I., Fedorova, O., Olinares, P.D.B., Chandanani, J., Banerjee, A., Choi, Y.J., Molina, H., Chait, B.T., Darst, S.A., Campbell, E.A.(2023) Mol Cell 83: 3921-3930.e7
- PubMed: 37890482
- DOI: https://doi.org/10.1016/j.molcel.2023.10.001
- Primary Citation of Related Structures:
8SQ9, 8SQJ, 8SQK - PubMed Abstract:
The enzymatic activity of the SARS-CoV-2 nidovirus RdRp-associated nucleotidyltransferase (NiRAN) domain is essential for viral propagation, with three distinct activities associated with modification of the nsp9 N terminus, NMPylation, RNAylation, and deRNAylation/capping via a GDP-polyribonucleotidyltransferase reaction. The latter two activities comprise an unconventional mechanism for initiating viral RNA 5' cap formation, while the role of NMPylation is unclear. The structural mechanisms for these diverse enzymatic activities have not been properly delineated. Here, we determine high-resolution cryoelectron microscopy (cryo-EM) structures of catalytic intermediates for the NMPylation and deRNAylation/capping reactions, revealing diverse nucleotide binding poses and divalent metal ion coordination sites to promote its repertoire of activities. The deRNAylation/capping structure explains why GDP is a preferred substrate for the capping reaction over GTP. Altogether, these findings enhance our understanding of the promiscuous coronaviral NiRAN domain, a therapeutic target, and provide an accurate structural platform for drug development.
Organizational Affiliation:
Laboratory of Molecular Biophysics, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA.