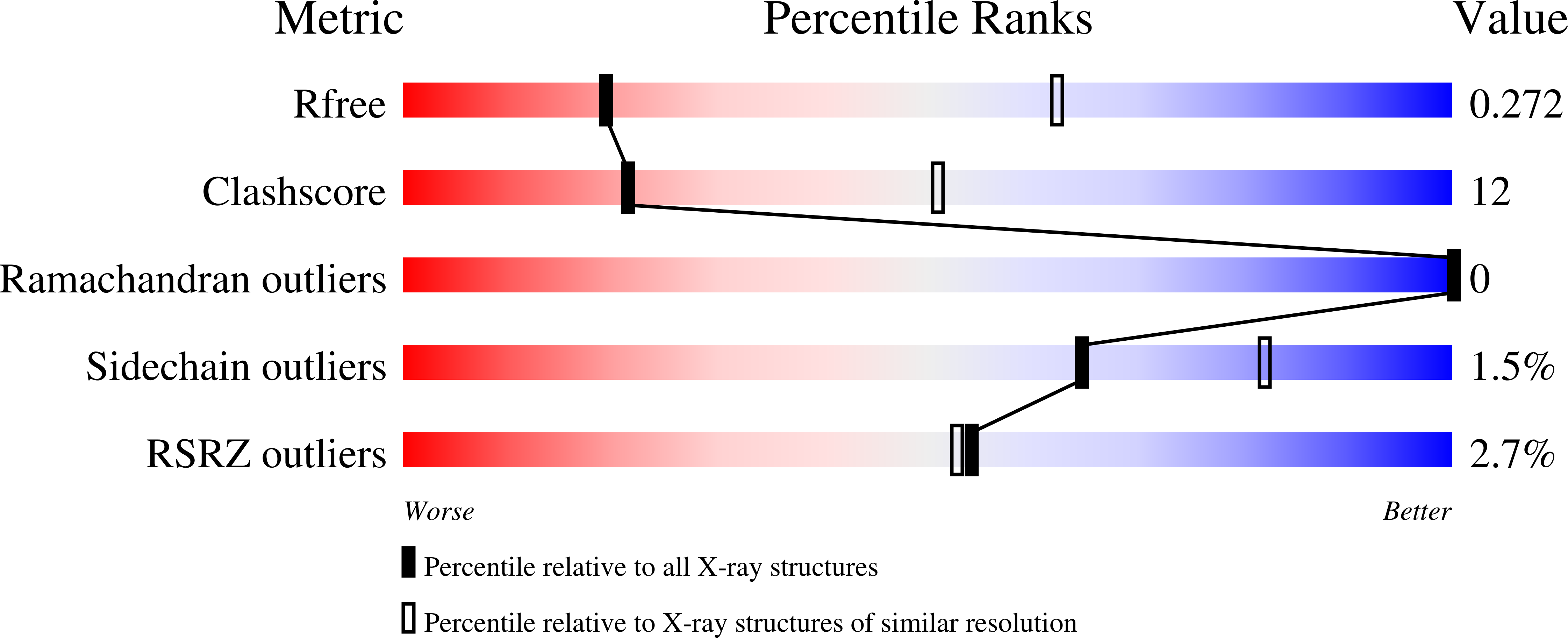
Identifying the molecular basis of Laminin N-terminal domain Ca 2+ binding using a hybrid approach.
Legare, S., Heide, F., Gabir, H., Rafiei, F., Meier, M., Padilla-Meier, G.P., Koch, M., Stetefeld, J.(2024) Biophys J 123: 2422-2430
- PubMed: 38851889
- DOI: https://doi.org/10.1016/j.bpj.2024.06.005
- Primary Citation of Related Structures:
8SNP - PubMed Abstract:
Ca 2+ is a highly abundant ion involved in numerous biological processes, particularly in multicellular eukaryotic organisms where it exerts many of these functions through interactions with Ca 2+ binding proteins. The laminin N-terminal (LN) domain is found in members of the laminin and netrin protein families where it plays a critical role in the function of these proteins. The LN domain of laminins and netrins is a Ca 2+ binding domain and in many cases requires Ca 2+ to perform its biological function. Here, we conduct a detailed examination of the molecular basis of the LN domain Ca 2+ interaction combining structural, computational, bioinformatics, and biophysical techniques. By combining computational and bioinformatic techniques with x-ray crystallography we explore the molecular basis of the LN domain Ca 2+ interaction and identify a conserved sequence present in Ca 2+ binding LN domains. These findings enable a sequence-based prediction of LN domain Ca 2+ binding ability. We use thermal shift assays and isothermal titration calorimetry to explore the biophysical properties of the LN domain Ca 2+ interaction. We show that the netrin-1 LN domain exhibits a high affinity and specificity for Ca 2+ , which structurally stabilizes the LN domain. This study elucidates the molecular foundation of the LN domain Ca 2+ binding interaction and provides a detailed functional characterization of this essential interaction, advancing our understanding of protein-Ca 2+ dynamics within the context of the LN domain.
Organizational Affiliation:
Department of Chemistry, University of Manitoba, Winnipeg, Manitoba, Canada. Electronic address: legares@myumanitoba.ca.