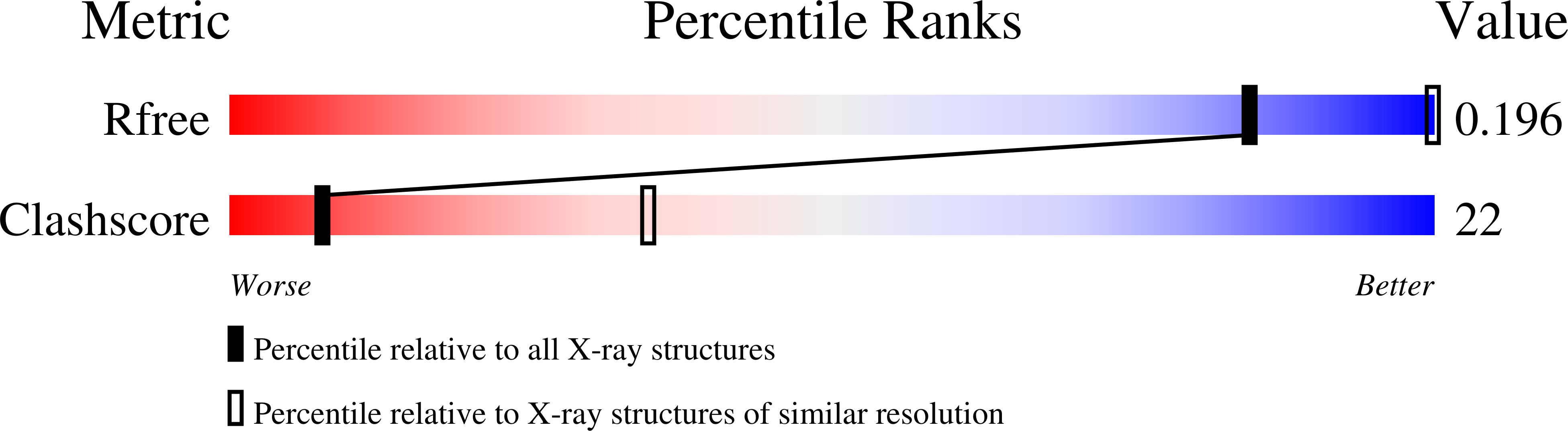Engineering tertiary chirality in helical biopolymers.
Janowski, J., Pham, V.A.B., Vecchioni, S., Woloszyn, K., Lu, B., Zou, Y., Erkalo, B., Perren, L., Rueb, J., Madnick, J., Mao, C., Saito, M., Ohayon, Y.P., Jonoska, N., Sha, R.(2024) Proc Natl Acad Sci U S A 121: e2321992121-e2321992121
- PubMed: 38684000
- DOI: https://doi.org/10.1073/pnas.2321992121
- Primary Citation of Related Structures:
8SJM, 8SJN, 8SJO, 8SJP, 8SJQ, 8SJR, 8SJS, 8SJT, 8SJU, 8SJV, 8SJW, 8SL5, 8SZ5 - PubMed Abstract:
Tertiary chirality describes the handedness of supramolecular assemblies and relies not only on the primary and secondary structures of the building blocks but also on topological driving forces that have been sparsely characterized. Helical biopolymers, especially DNA, have been extensively investigated as they possess intrinsic chirality that determines the optical, mechanical, and physical properties of the ensuing material. Here, we employ the DNA tensegrity triangle as a model system to locate the tipping points in chirality inversion at the tertiary level by X-ray diffraction. We engineer tensegrity triangle crystals with incremental rotational steps between immobile junctions from 3 to 28 base pairs (bp). We construct a mathematical model that accurately predicts and explains the molecular configurations in both this work and previous studies. Our design framework is extendable to other supramolecular assemblies of helical biopolymers and can be used in the design of chiral nanomaterials, optically active molecules, and mesoporous frameworks, all of which are of interest to physical, biological, and chemical nanoscience.
Organizational Affiliation:
Department of Chemistry, New York University, New York, NY 10003.