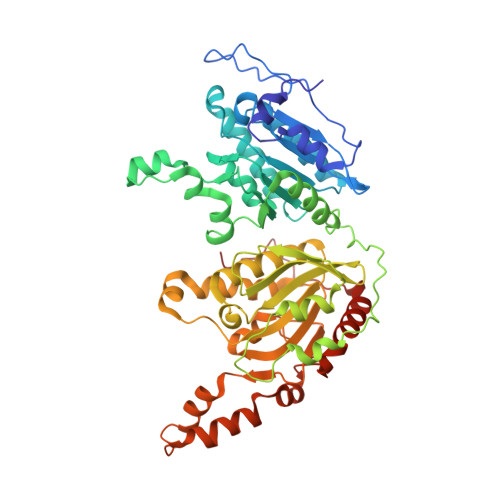
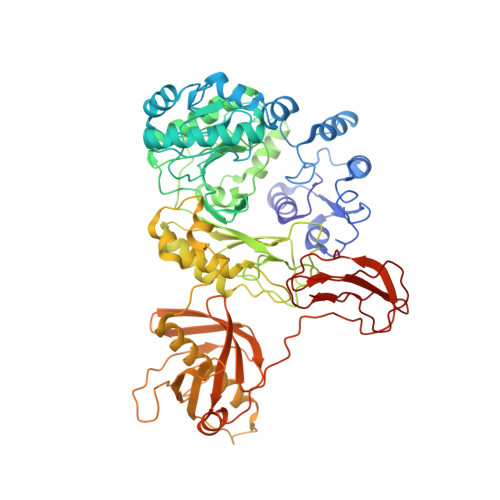
CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics.
Lee, J.K.J., Liu, Y.T., Hu, J.J., Aphasizheva, I., Aphasizhev, R., Zhou, Z.H.(2023) J Struct Biol X 7: 100088-100088
- PubMed: 37128595
- DOI: https://doi.org/10.1016/j.yjsbx.2023.100088
- Primary Citation of Related Structures:
8SGX, 8SGY, 8SGZ - PubMed Abstract:
Propionyl-CoA carboxylase (PCC) is a multienzyme complex consisting of up to six α-subunits and six β-subunits. Belonging to a metabolic pathway converging on the citric acid cycle, it is present in most forms of life and irregularities in its assembly lead to serious illness in humans, known as propionic acidemia. Here, we report the cryogenic electron microscopy (cryoEM) structures and assembly of different oligomeric isomers of endogenous PCC from the parasitic protozoan Leishmania tarentolae (LtPCC). These structures and their statistical distribution reveal the mechanics of PCC assembly and disassembly at equilibrium. We show that, in solution, endogenous LtPCC β-subunits form stable homohexamers, to which different numbers of α-subunits attach. Sorting LtPCC particles into seven classes (i.e., oligomeric formulae α 0 β 6 , α 1 β 6 , α 2 β 6 , α 3 β 6 , α 4 β 6 , α 5 β 6 , α 6 β 6 ) enables formulation of a model for PCC assembly. Our results suggest how multimerization regulates PCC enzymatic activity and showcase the utility of cryoEM in revealing the statistical mechanics of reaction pathways.
Organizational Affiliation:
Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles (UCLA), Los Angeles, CA 90095, USA.