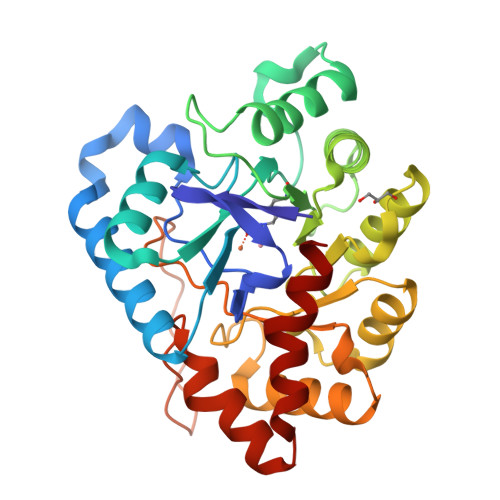
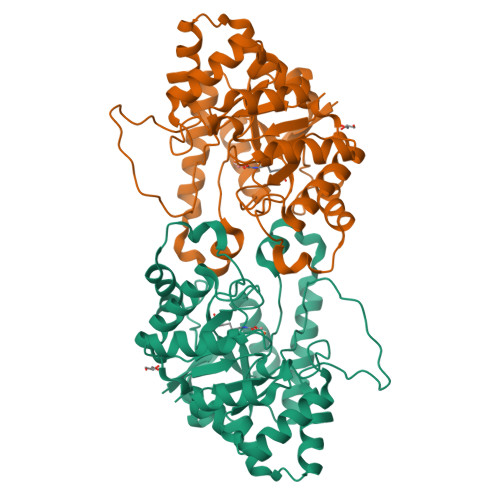
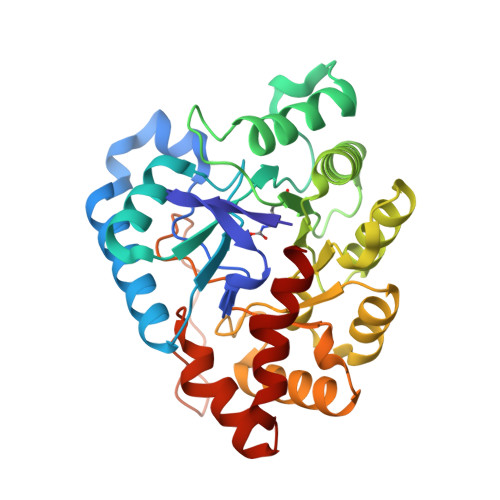
Changes in Active Site Loop Conformation Relate to the Transition toward a Novel Enzymatic Activity.
Jacquet, P., Billot, R., Shimon, A., Hoekstra, N., Bergonzi, C., Jenks, A., Chabriere, E., Daude, D., Elias, M.H.(2024) JACS Au 4: 1941-1953
- PubMed: 38818068
- DOI: https://doi.org/10.1021/jacsau.4c00179
- Primary Citation of Related Structures:
8SF2, 8SF9, 8SFA, 8SFB, 8SFC, 8SFD, 8SFK, 8SFM - PubMed Abstract:
Enzymatic promiscuity, the ability of enzymes to catalyze multiple, distinct chemical reactions, has been well documented and is hypothesized to be a major driver of the emergence of new enzymatic functions. Yet, the molecular mechanisms involved in the transition from one activity to another remain debated and elusive. Here, we evaluated the redesign of the active site binding cleft of lactonase Sso Pox using structure-based design and combinatorial libraries. We created variants with largely improved catalytic abilities against phosphotriesters, the best ones being >1000-fold better compared to the wild-type enzyme. The observed shifts in activity specificity are large, and some variants completely lost their initial activity. The selected combinations of mutations have considerably reshaped the active site cavity via side chain changes but mostly through large rearrangements of the active site loops and changes to their conformations, as revealed by a suite of crystal structures. This suggests that a specific active site loop configuration is critical to the lactonase activity. Interestingly, analysis of high-resolution structures hints at the potential role of conformational sampling and its directionality in defining the enzyme activity profile.
Organizational Affiliation:
Gene&GreenTK, 19-21 Bd Jean Moulin, Marseille 13005, France.