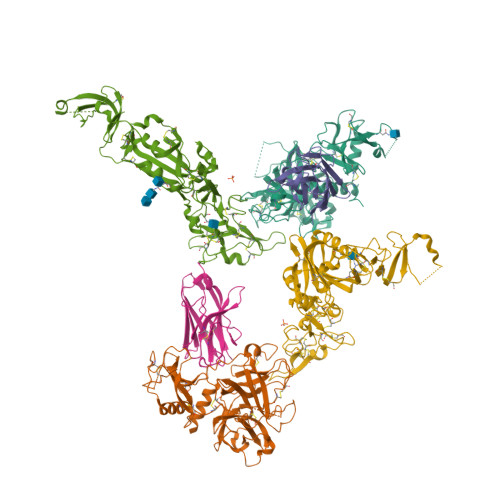
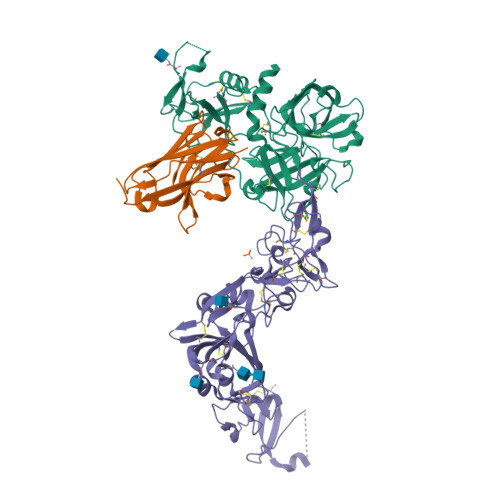
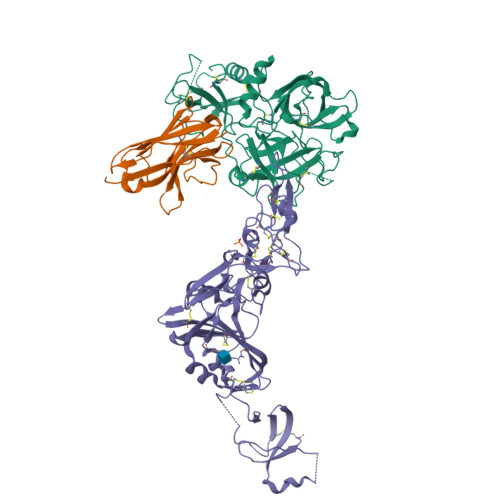
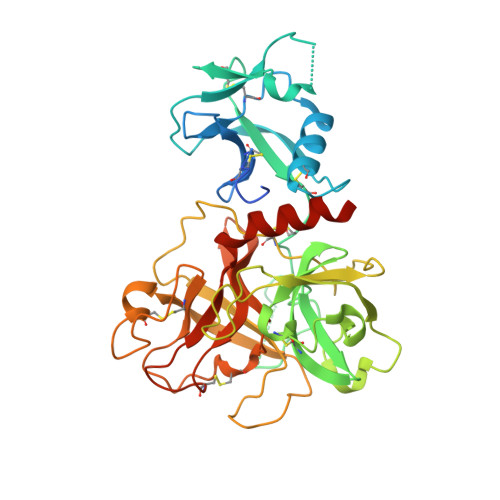
Structural basis of TMPRSS2 zymogen activation and recognition by the HKU1 seasonal coronavirus.
Fernandez, I., Saunders, N., Duquerroy, S., Bolland, W.H., Arbabian, A., Baquero, E., Blanc, C., Lafaye, P., Haouz, A., Buchrieser, J., Schwartz, O., Rey, F.A.(2024) Cell 187: 4246-4260.e16
- PubMed: 38964326
- DOI: https://doi.org/10.1016/j.cell.2024.06.007
- Primary Citation of Related Structures:
8S0L, 8S0M, 8S0N - PubMed Abstract:
The human seasonal coronavirus HKU1-CoV, which causes common colds worldwide, relies on the sequential binding to surface glycans and transmembrane serine protease 2 (TMPRSS2) for entry into target cells. TMPRSS2 is synthesized as a zymogen that undergoes autolytic activation to process its substrates. Several respiratory viruses, in particular coronaviruses, use TMPRSS2 for proteolytic priming of their surface spike protein to drive membrane fusion upon receptor binding. We describe the crystal structure of the HKU1-CoV receptor binding domain in complex with TMPRSS2, showing that it recognizes residues lining the catalytic groove. Combined mutagenesis of interface residues and comparison across species highlight positions 417 and 469 as determinants of HKU1-CoV host tropism. The structure of a receptor-blocking nanobody in complex with zymogen or activated TMPRSS2 further provides the structural basis of TMPRSS2 activating conformational change, which alters loops recognized by HKU1-CoV and dramatically increases binding affinity.
Organizational Affiliation:
Institut Pasteur, Université de Paris Cité, CNRS UMR 3569, Structural Virology Unit, 75015 Paris, France.