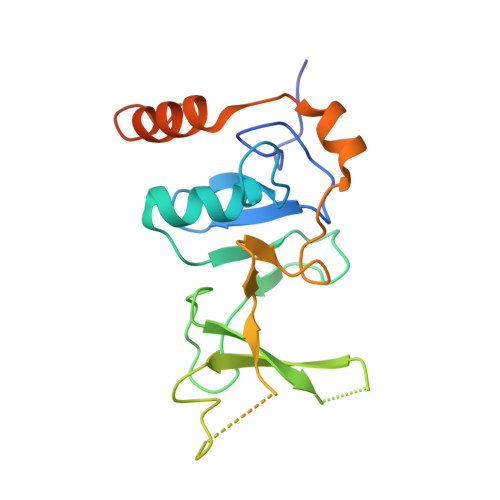
UPF1 helicase orchestrates mutually exclusive interactions with the SMG6 endonuclease and UPF2.
Langer, L.M., Kurscheidt, K., Basquin, J., Bonneau, F., Iermak, I., Basquin, C., Conti, E.(2024) Nucleic Acids Res 52: 6036-6048
- PubMed: 38709891
- DOI: https://doi.org/10.1093/nar/gkae323
- Primary Citation of Related Structures:
8RXB - PubMed Abstract:
Nonsense-mediated mRNA decay (NMD) is a conserved co-translational mRNA surveillance and turnover pathway across eukaryotes. NMD has a central role in degrading defective mRNAs and also regulates the stability of a significant portion of the transcriptome. The pathway is organized around UPF1, an RNA helicase that can interact with several NMD-specific factors. In human cells, degradation of the targeted mRNAs begins with a cleavage event that requires the recruitment of the SMG6 endonuclease to UPF1. Previous studies have identified functional links between SMG6 and UPF1, but the underlying molecular mechanisms have remained elusive. Here, we used mass spectrometry, structural biology and biochemical approaches to identify and characterize a conserved short linear motif in SMG6 that interacts with the cysteine/histidine-rich (CH) domain of UPF1. Unexpectedly, we found that the UPF1-SMG6 interaction is precluded when the UPF1 CH domain is engaged with another NMD factor, UPF2. Based on cryo-EM data, we propose that the formation of distinct SMG6-containing and UPF2-containing NMD complexes may be dictated by different conformational states connected to the RNA-binding status of UPF1. Our findings rationalize a key event in metazoan NMD and advance our understanding of mechanisms regulating activity and guiding substrate recognition by the SMG6 endonuclease.
Organizational Affiliation:
Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Martinsried/Munich D-82152, Germany.