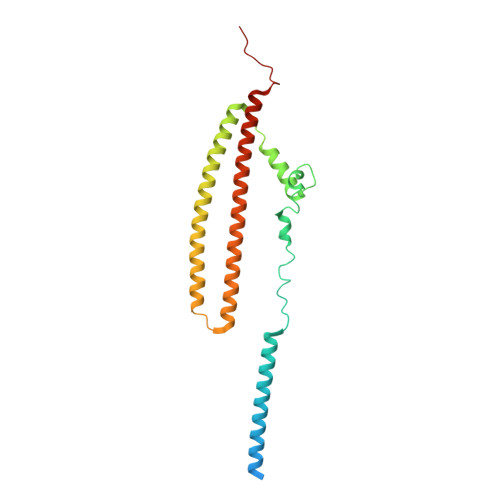
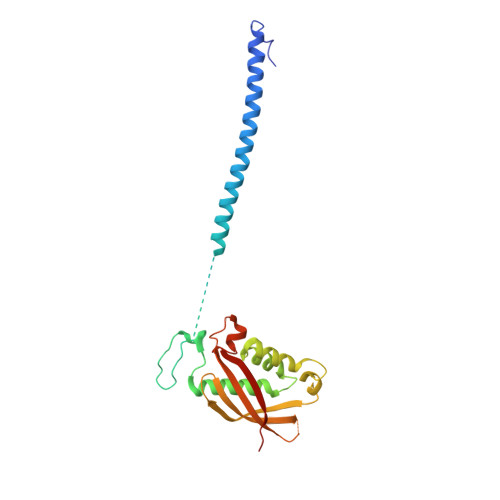
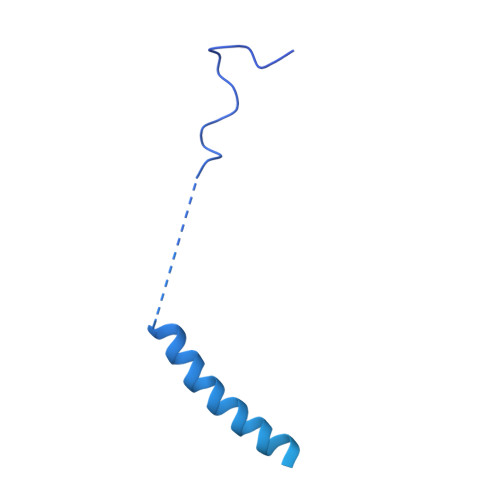
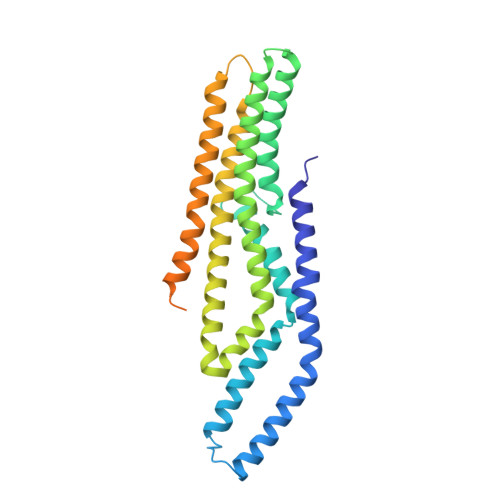
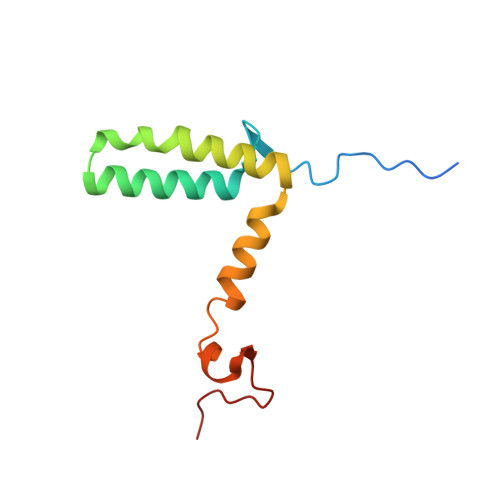
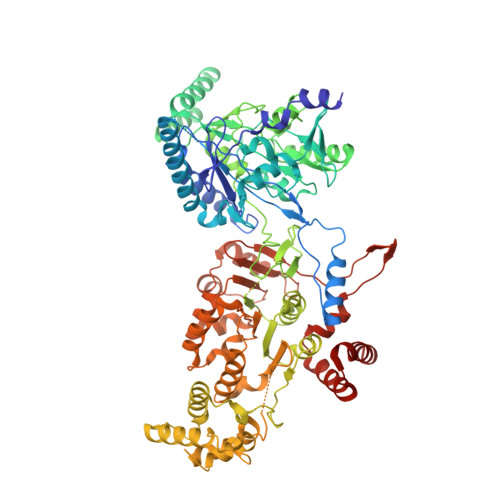
Cryo-EM structure of a conjugative type IV secretion system suggests a molecular switch regulating pilus biogenesis.
Mace, K., Waksman, G.(2024) EMBO J
- PubMed: 38886579
- DOI: https://doi.org/10.1038/s44318-024-00135-z
- Primary Citation of Related Structures:
8RT4, 8RT5, 8RT6, 8RT7, 8RT8, 8RT9, 8RTA, 8RTB, 8RTD - PubMed Abstract:
Conjugative type IV secretion systems (T4SS) mediate bacterial conjugation, a process that enables the unidirectional exchange of genetic materials between a donor and a recipient bacterial cell. Bacterial conjugation is the primary means by which antibiotic resistance genes spread among bacterial populations (Barlow 2009; Virolle et al, 2020). Conjugative T4SSs form pili: long extracellular filaments that connect with recipient cells. Previously, we solved the cryo-electron microscopy (cryo-EM) structure of a conjugative T4SS. In this article, based on additional data, we present a more complete T4SS cryo-EM structure than that published earlier. Novel structural features include details of the mismatch symmetry within the OMCC, the presence of a fourth VirB8 subunit in the asymmetric unit of both the arches and the inner membrane complex (IMC), and a hydrophobic VirB5 tip in the distal end of the stalk. Additionally, we provide previously undescribed structural insights into the protein VirB10 and identify a novel regulation mechanism of T4SS-mediated pilus biogenesis by this protein, that we believe is a key checkpoint for this process.
Organizational Affiliation:
Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck College, Malet Street, London, WC1E 7HX, UK. kevin.mace@univ-rennes.fr.