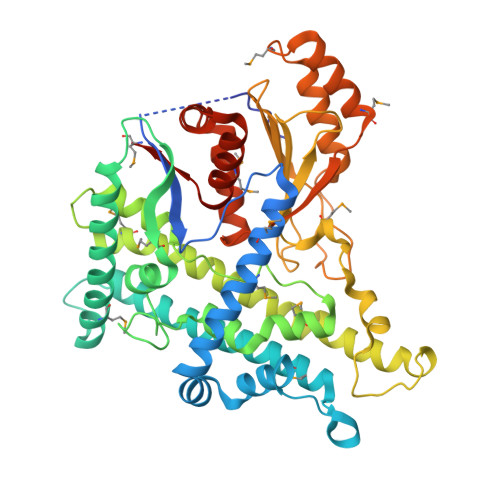
The effect of pH on the structure of Bluetongue virus VP5.
Zhang, H., El Omari, K., Sutton, G., Stuart, D.I.(2024) J Gen Virol 105
- PubMed: 39163113
- DOI: https://doi.org/10.1099/jgv.0.002018
- Primary Citation of Related Structures:
8RT0, 8RT1 - PubMed Abstract:
The unenveloped Bluetongue virus capsid comprises several structural layers, the inner two comprising a core, which assembles before addition of the outer proteins, VP2 and VP5. Two symmetric trimers of VP5 fit like pegs into two distinct pits on the core and undergo pH conformational changes in the context of the virus, associated with cell entry. Here we show that in isolation VP5 alone undergoes essentially the same changes with pH and confirm a helical transition, indicating that VP5 is a motor during cell entry. In the absence of VP5 the two pits on the core differ from each other, presumably due to the asymmetric underlying structure of VP3, the innermost capsid protein. On insertion of VP5 these pits become closely similar and remain similar at low pH whilst VP5 is present. This natural asymmetry presumably destabilises the attachment of VP5, facilitating ejection upon low pH, membrane penetration and cell entry.
Organizational Affiliation:
Division of Structural Biology, University of Oxford, Centre for Human Genetics, Oxford, UK.