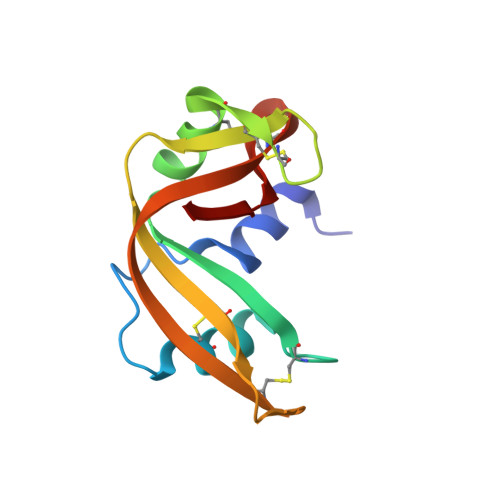
Crystal structure of two covalent nucleoside derivatives of ribonuclease A.
Nachman, J., Miller, M., Gilliland, G.L., Carty, R., Pincus, M., Wlodawer, A.(1990) Biochemistry 29: 928-937
- PubMed: 2340284
- DOI: https://doi.org/10.1021/bi00456a012
- Primary Citation of Related Structures:
8RSA, 9RSA - PubMed Abstract:
Crystal structures of two forms of ribonuclease A with deoxynucleosides covalently bound to respectively His 12 and His 119 have been solved. One form, T-H12-RNase, has a deoxythymidine bound to N epsilon 2 of His 12, while the other one, U-H119-RNase, has a deoxyuridine bound to N delta 1 of His 119. The two crystal forms are nearly isomorphous, with two molecules in the asymmetric unit. However, the modified ribonucleases differ both in their enzymatic activities and in the conformation of the catalytic site and of the deoxynucleoside-histidine moiety. T-H12-RNase is characterized by complete loss of enzymatic activity; in this form the deoxynucleoside completely blocks the catalytic site and forms intramolecular contacts with residues associated with both the B1 and B2 sites. U-H119-RNase retains 1% of the activity of the unmodified enzyme, and in this form His 119 adopts a different orientation, corresponding to the alternate conformation reported for this residue; the deoxynucleoside-histidine moiety points out of the active site and does not form any contacts with the rest of the protein, thus allowing partial access to the catalytic site. On the basis of these structures, we propose possible mechanisms for the reactions of bromoacetamido nucleosides with ribonuclease A.
Organizational Affiliation:
BRI Basic Research Program, NCI-Frederick Cancer Research Facility, Maryland 21701.