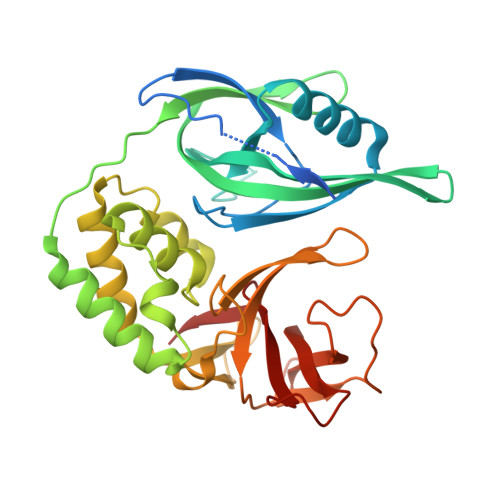
Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders
Kroupova, A., Spiteri, V., Furihata, H., Darren, D., Ramachandran, S., Rutter, Z., Chakraborti, S., Haubrich, K., Pethe, J., Gonzales, D., Wijaya, A., Rodriguez-Rios, M., Lynch, D., Farnaby, W., Nakasone, M., Zollman, D., Ciulli, A.To be published.