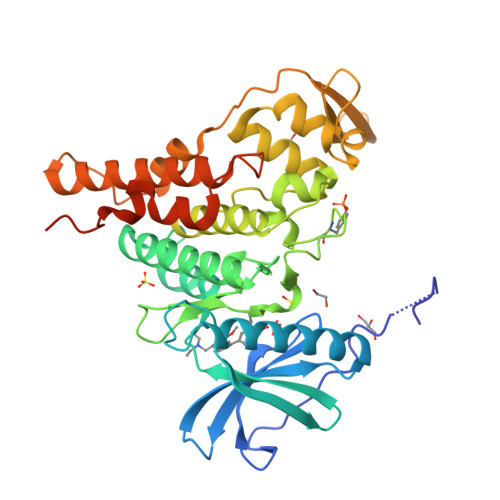
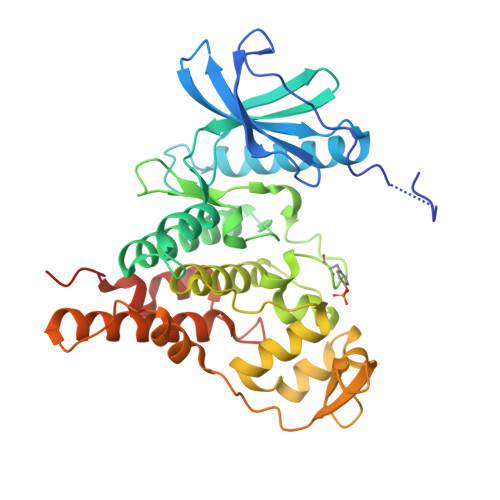
Targeting Pf CLK3 with Covalent Inhibitors: A Novel Strategy for Malaria Treatment.
Brettell, S.B., Janha, O., Begen, A., Cann, G., Sharma, S., Olaniyan, N., Yelland, T., Hole, A.J., Alam, B., Mayville, E., Gillespie, R., Capper, M., Fidock, D.A., Milligan, G., Clarke, D.J., Tobin, A.B., Jamieson, A.G.(2024) J Med Chem 67: 18895-18910
- PubMed: 39441986
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01300
- Primary Citation of Related Structures:
8RPC - PubMed Abstract:
Malaria still causes over 600,000 deaths annually, with rising resistance to frontline drugs by Plasmodium falciparum increasing this number each year. New medicines with novel mechanisms of action are, therefore, urgently needed. In this work, we solved the cocrystal structure of the essential malarial kinase Pf CLK3 with the reversible inhibitor TCMDC-135051 ( 1 ), enabling the design of covalent inhibitors targeting a unique cysteine residue (Cys368) poorly conserved in the human kinome. Chloroacetamide 4 shows nanomolar potency and covalent inhibition in both recombinant protein and P. falciparum assays. Efficacy in parasites persisted after a 6 h washout, indicating an extended duration of action. Additionally, 4 showed improved kinase selectivity and a high selectivity index against HepG2 cells, with a low propensity for resistance (log MIR > 8.1). To our knowledge, compound 4 is the first covalent inhibitor of a malarial kinase, offering promising potential as a lead for a single-dose malaria cure.
Organizational Affiliation:
School of Chemistry, The Advanced Research Centre, University of Glasgow, 11 Chapel Lane, Glasgow G11 6EW, U.K.