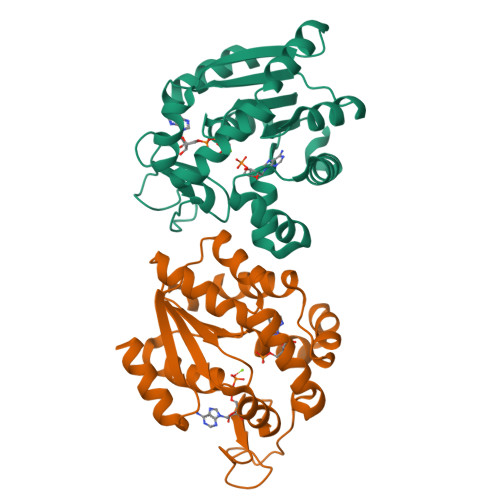
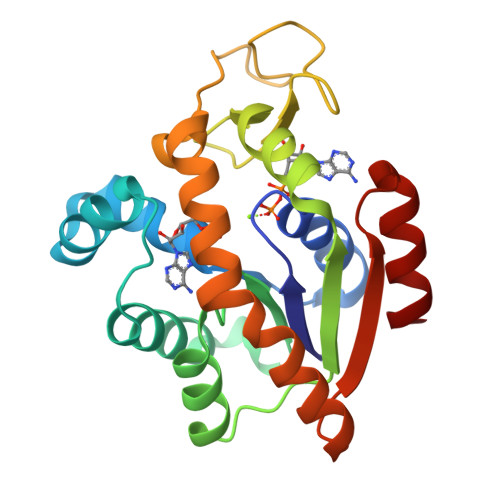
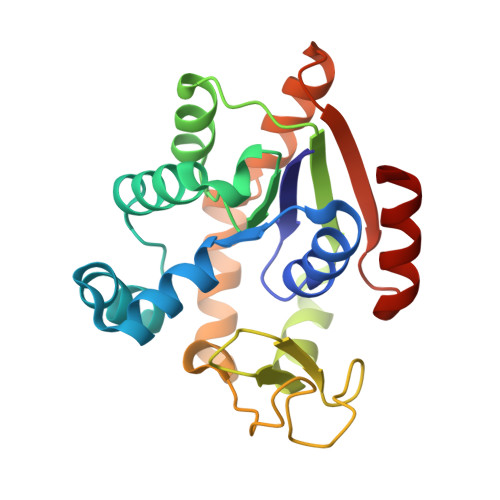
Magnesium induced structural reorganization in the active site of adenylate kinase.
Nam, K., Thodika, A.R.A., Tischlik, S., Phoeurk, C., Nagy, T.M., Schierholz, L., Aden, J., Rogne, P., Drescher, M., Sauer-Eriksson, A.E., Wolf-Watz, M.(2024) Sci Adv 10: eado5504-eado5504
- PubMed: 39121211
- DOI: https://doi.org/10.1126/sciadv.ado5504
- Primary Citation of Related Structures:
8RJ4, 8RJ6, 8RJ9 - PubMed Abstract:
Phosphoryl transfer is a fundamental reaction in cellular signaling and metabolism that requires Mg 2+ as an essential cofactor. While the primary function of Mg 2+ is electrostatic activation of substrates, such as ATP, the full spectrum of catalytic mechanisms exerted by Mg 2+ is not known. In this study, we integrate structural biology methods, molecular dynamic (MD) simulations, phylogeny, and enzymology assays to provide molecular insights into Mg 2+ -dependent structural reorganization in the active site of the metabolic enzyme adenylate kinase. Our results demonstrate that Mg 2+ induces a conformational rearrangement of the substrates (ATP and ADP), resulting in a 30° adjustment of the angle essential for reversible phosphoryl transfer, thereby optimizing it for catalysis. MD simulations revealed transitions between conformational substates that link the fluctuation of the angle to large-scale enzyme dynamics. The findings contribute detailed insight into Mg 2+ activation of enzymes and may be relevant for reversible and irreversible phosphoryl transfer reactions.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Texas at Arlington, Arlington, TX 76019, USA.