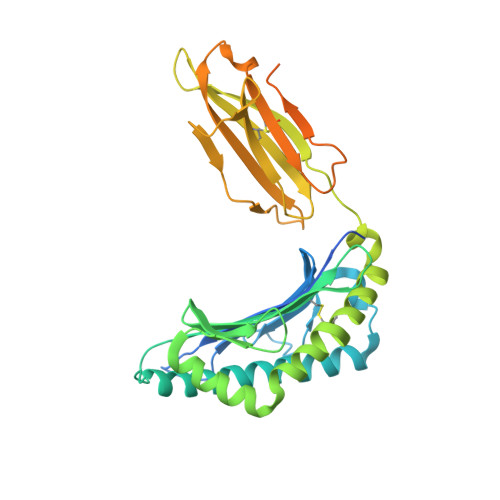
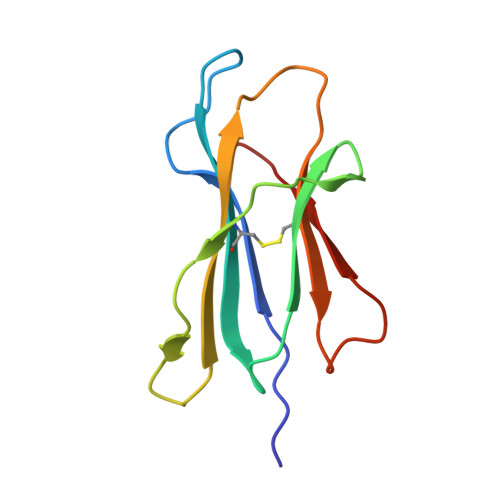
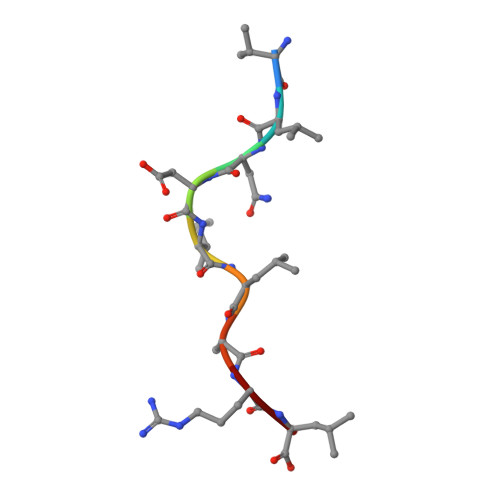
The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Ahn, Y.M., Maddumage, J.C., Grant, E.J., Chatzileontiadou, D.S.M., Perera, W.W.J.G., Baker, B.M., Szeto, C., Gras, S.(2024) Curr Res Struct Biol 7: 100148-100148
- PubMed: 38742159
- DOI: https://doi.org/10.1016/j.crstbi.2024.100148
- Primary Citation of Related Structures:
8RBU, 8RBV, 8RCV, 8REF, 8RH6, 8RHQ - PubMed Abstract:
CD8 + T cells are crucial for viral elimination and recovery from viral infection. Nonetheless, the current understanding of the T cell response to SARS-CoV-2 at the antigen level remains limited. The Spike protein is an external structural protein that is prone to mutations, threatening the efficacy of current vaccines. Therefore, we have characterised the immune response towards the immunogenic Spike-derived peptide (S 976-984 , VLNDILSRL), restricted to the HLA-A*02:01 molecule, which is mutated in both Alpha (S982A) and Omicron BA.1 (L981F) variants of concern. We determined that the mutation in the Alpha variant (S982A) impacted both the stability and conformation of the peptide, bound to HLA-A*02:01, in comparison to the original S 976-984 . We identified a longer and overlapping immunogenic peptide (S 975-984 , SVLNDILSRL) that could be presented by HLA-A*02:01, HLA-A*11:01 and HLA-B*13:01 allomorphs. We showed that S975-specific CD8 + T cells were weakly cross-reactive to the mutant peptides despite their similar conformations when presented by HLA-A*11:01. Altogether, our results show that the impact of SARS-CoV-2 mutations on peptide presentation is HLA allomorph-specific, and that post vaccination there are T cells able to react and cross-react towards the variant of concern peptides.
- Infection & Immunity Program, La Trobe Institute for Molecular Science (LIMS), La Trobe University, Bundoora, Victoria, Australia.
Organizational Affiliation: