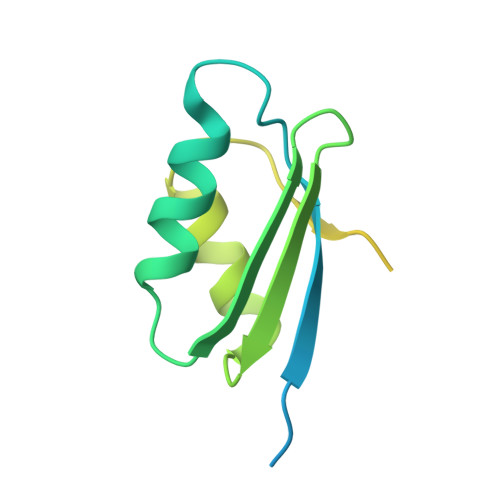
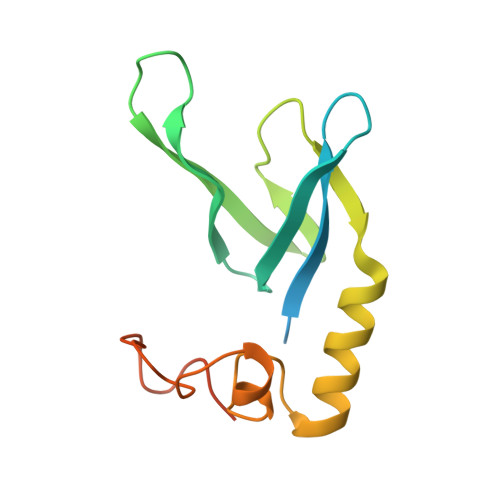
Bioengineering a plant NLR immune receptor with a robust binding interface toward a conserved fungal pathogen effector.
Zdrzalek, R., Xi, Y., Langner, T., Bentham, A.R., Petit-Houdenot, Y., De la Concepcion, J.C., Harant, A., Shimizu, M., Were, V., Talbot, N.J., Terauchi, R., Kamoun, S., Banfield, M.J.(2024) Proc Natl Acad Sci U S A 121: e2402872121-e2402872121
- PubMed: 38968126
- DOI: https://doi.org/10.1073/pnas.2402872121
- Primary Citation of Related Structures:
8R7A, 8R7D - PubMed Abstract:
Bioengineering of plant immune receptors has emerged as a key strategy for generating novel disease resistance traits to counteract the expanding threat of plant pathogens to global food security. However, current approaches are limited by rapid evolution of plant pathogens in the field and may lack durability when deployed. Here, we show that the rice nucleotide-binding, leucine-rich repeat (NLR) immune receptor Pik-1 can be engineered to respond to a conserved family of effectors from the multihost blast fungus pathogen Magnaporthe oryzae . We switched the effector binding and response profile of the Pik NLR from its cognate rice blast effector AVR-Pik to the host-determining factor pathogenicity toward weeping lovegrass 2 (Pwl2) by installing a putative host target, OsHIPP43, in place of the native integrated heavy metal-associated domain (generating Pikm-1 OsHIPP43 ). This chimeric receptor also responded to other PWL alleles from diverse blast isolates. The crystal structure of the Pwl2/OsHIPP43 complex revealed a multifaceted, robust interface that cannot be easily disrupted by mutagenesis, and may therefore provide durable, broad resistance to blast isolates carrying PWL effectors in the field. Our findings highlight how the host targets of pathogen effectors can be used to bioengineer recognition specificities that have more robust properties compared to naturally evolved disease resistance genes.
Organizational Affiliation:
Department of Biochemistry and Metabolism, John Innes Centre, Norwich NR4 7UH, United Kingdom.