Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
Czestkowski, W., Krzeminski, L., Piotrowicz, M.C., Mazur, M., Pluta, E., Andryianau, G., Koralewski, R., Matyszewski, K., Olejniczak, S., Kowalski, M., Lisiecka, K., Koziel, R., Piwowar, K., Papiernik, D., Nowotny, M., Napiorkowska-Gromadzka, A., Nowak, E., Niedzialek, D., Wieczorek, G., Siwinska, A., Rejczak, T., Jedrzejczak, K., Mulewski, K., Olczak, J., Zaslona, Z., Golebiowski, A., Drzewicka, K., Bartoszewicz, A.(2024) J Med Chem 67: 3959-3985
- PubMed: 38427954
- DOI: https://doi.org/10.1021/acs.jmedchem.3c02255
- Primary Citation of Related Structures:
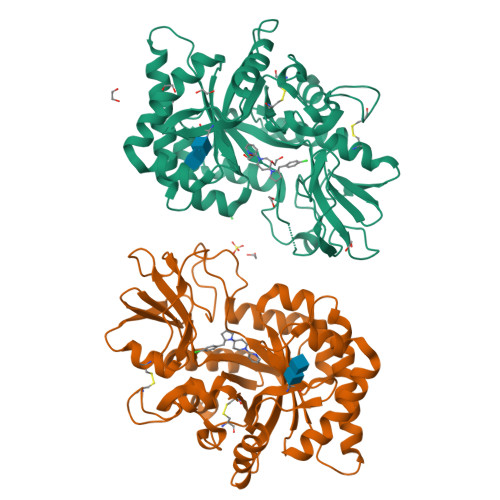
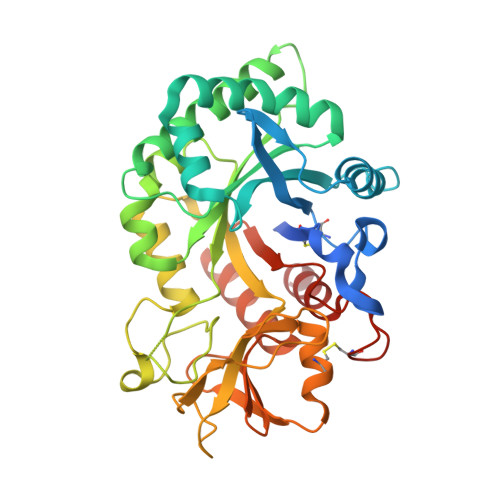
8R41, 8R42, 8R4X - PubMed Abstract:
Chitinase-3-like-1 (CHI3L1), also known as YKL-40, is a glycoprotein linked to inflammation, fibrosis, and cancer. This study explored CHI3L1's interactions with various oligosaccharides using microscale thermophoresis (MST) and AlphaScreen (AS). These investigations guided the development of high-throughput screening assays to assess interference of small molecules in binding between CHI3L1 and biotinylated small molecules or heparan sulfate-based probes. Small molecule binders of YKL-40 were identified in our chitotriosidase inhibitors library with MST and confirmed through X-ray crystallography. Based on cocrystal structures of potent hit compounds with CHI3L1, small molecule probes 19 and 20 were designed for an AS assay. Structure-based optimization led to compounds 30 and 31 with nanomolar activities and drug-like properties. Additionally, an orthogonal AS assay using biotinylated heparan sulfate as a probe was developed. The compounds' affinity showed a significant correlation in both assays. These screening tools and compounds offer novel avenues for investigating the role of CHI3L1.
Organizational Affiliation:
Molecure S.A., Żwirki I Wigury 101, Warsaw 02-089, Poland.