Structural and functional analysis of the minimal orthomyxovirus-like polymerase of Tilapia Lake Virus from the highly diverged Amnoonviridae family.
Arragain, B., Pelosse, M., Thompson, A., Cusack, S.(2023) Nat Commun 14: 8145-8145
- PubMed: 38066000
- DOI: https://doi.org/10.1038/s41467-023-44044-x
- Primary Citation of Related Structures:
8PSN, 8PSO, 8PSQ, 8PSS, 8PSU, 8PSX, 8PSZ, 8PT2, 8PT6, 8PT7, 8PTH, 8PTJ, 8QZ8 - PubMed Abstract:
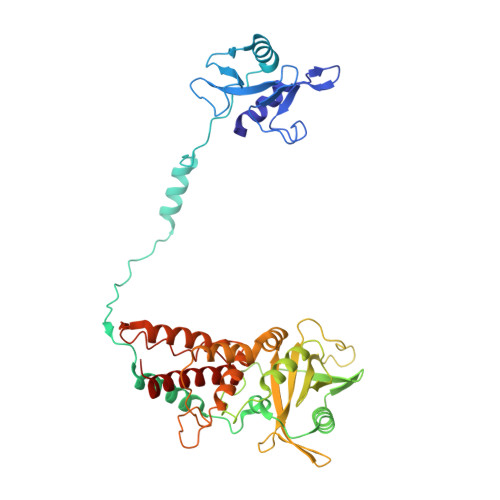
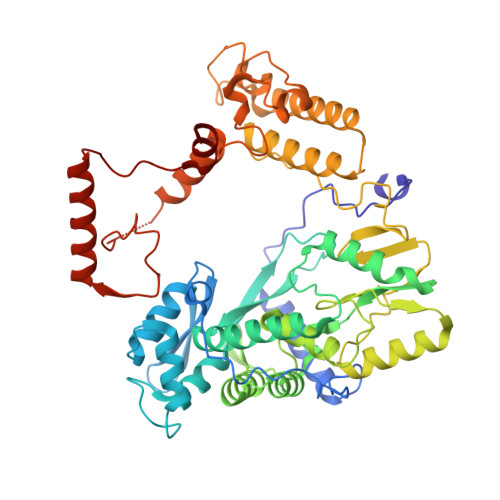
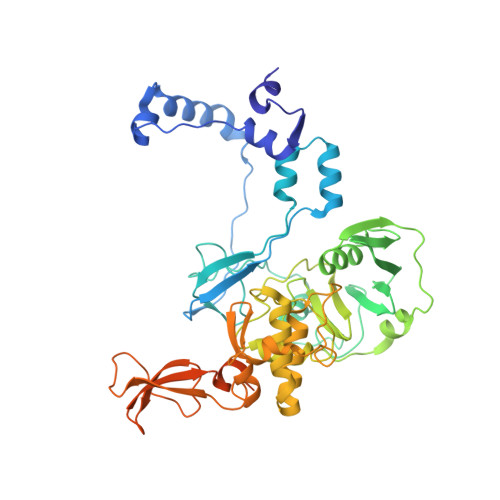
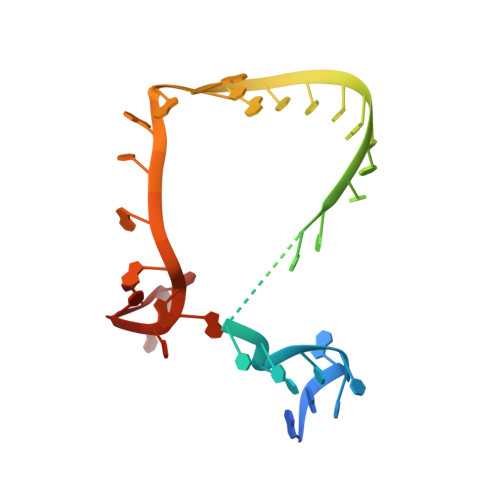
Tilapia Lake Virus (TiLV), a recently discovered pathogen of tilapia fish, belongs to the Amnoonviridae family from the Articulavirales order. Its ten genome segments have characteristic conserved ends and encode proteins with no known homologues, apart from the segment 1, which encodes an orthomyxo-like RNA-dependent-RNA polymerase core subunit. Here we show that segments 1-3 encode respectively the PB1, PB2 and PA-like subunits of an active heterotrimeric polymerase that maintains all domains found in the distantly related influenza polymerase, despite an unprecedented overall size reduction of 40%. Multiple high-resolution cryo-EM structures of TiLV polymerase in pre-initiation, initiation and active elongation states, show how it binds the vRNA and cRNA promoters and performs RNA synthesis, with both transcriptase and replicase configurations being characterised. However, the highly truncated endonuclease-like domain appears inactive and the putative cap-binding domain is autoinhibited, emphasising that many functional aspects of TiLV polymerase remain to be elucidated.
Organizational Affiliation:
European Molecular Biology Laboratory, 71 Avenue des Martyrs, CS 90181, 38042, Grenoble, Cedex 9, France.