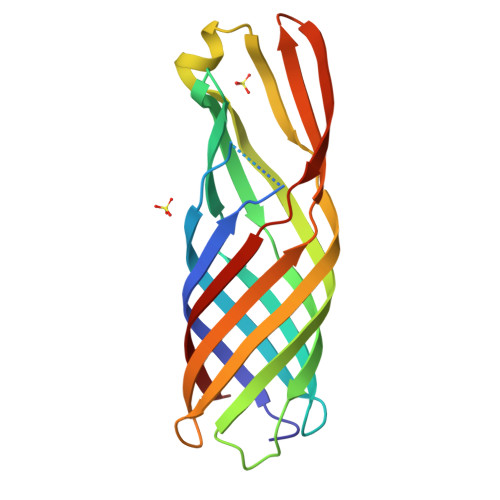
Structure of the outer membrane porin OmpW from the pervasive pathogen Klebsiella pneumoniae.
Seddon, C., Frankel, G., Beis, K.(2024) Acta Crystallogr F Struct Biol Commun 80: 22-27
- PubMed: 38206593
- DOI: https://doi.org/10.1107/S2053230X23010579
- Primary Citation of Related Structures:
8QXP - PubMed Abstract:
Conjugation is the process by which plasmids, including those that carry antibiotic-resistance genes, are mobilized from one bacterium (the donor) to another (the recipient). The conjugation efficiency of IncF-like plasmids relies on the formation of mating-pair stabilization via intimate interactions between outer membrane proteins on the donor (a plasmid-encoded TraN isoform) and recipient bacteria. Conjugation of the R100-1 plasmid into Escherichia coli and Klebsiella pneumoniae (KP) recipients relies on pairing between the plasmid-encoded TraNα in the donor and OmpW in the recipient. Here, the crystal structure of K. pneumoniae OmpW (OmpW KP ) is reported at 3.2 Å resolution. OmpW KP forms an eight-stranded β-barrel flanked by extracellular loops. The structures of E. coli OmpW (OmpW EC ) and OmpW KP show high conservation despite sequence variability in the extracellular loops.
Organizational Affiliation:
Department of Life Sciences, Imperial College, London, United Kingdom.