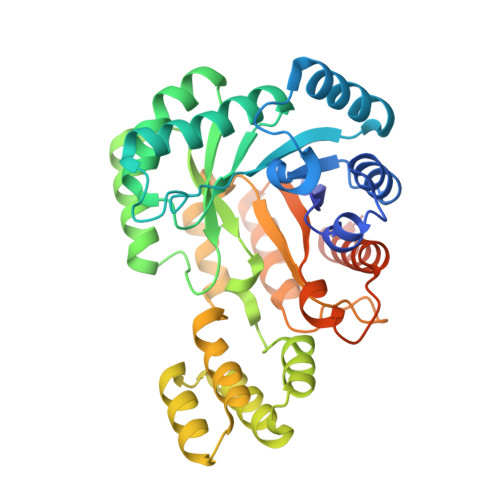
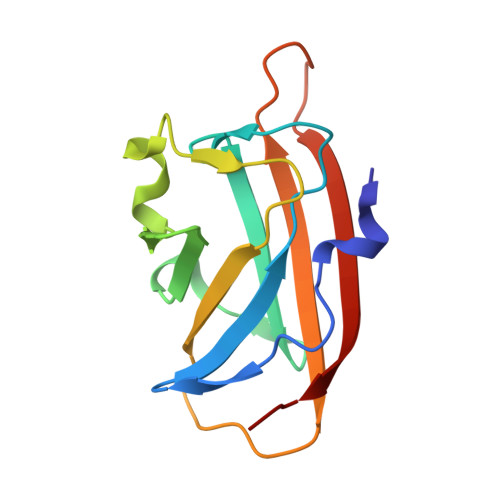
Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
Furlanetto, V., Kalyani, D.C., Kostelac, A., Puc, J., Haltrich, D., Hallberg, B.M., Divne, C.(2024) J Mol Biol 436: 168547-168547
- PubMed: 38508304
- DOI: https://doi.org/10.1016/j.jmb.2024.168547
- Primary Citation of Related Structures:
8QVC, 8QVD, 8QVE, 8UMC - PubMed Abstract:
Plant C-glycosylated aromatic polyketides are important for plant and animal health. These are specialized metabolites that perform functions both within the plant, and in interaction with soil or intestinal microbes. Despite the importance of these plant compounds, there is still limited knowledge of how they are metabolized. The Gram-positive aerobic soil bacterium Deinococcus aerius strain TR0125 and other Deinococcus species thrive in a wide range of harsh environments. In this work, we identified a C-glycoside deglycosylation gene cluster in the genome of D. aerius. The cluster includes three genes coding for a GMC-type oxidoreductase (DaCGO1) that oxidizes the glucosyl C3 position in aromatic C-glucosyl compounds, which in turn provides the substrate for the C-glycoside deglycosidase (DaCGD; composed of α+β subunits) that cleaves the glucosyl-aglycone C-C bond. Our results from size-exclusion chromatography, single particle cryo-electron microscopy and X-ray crystallography show that DaCGD is an α 2 β 2 heterotetramer, which represents a novel oligomeric state among bacterial CGDs. Importantly, the high-resolution X-ray structure of DaCGD provides valuable insights into the activation of the catalytic hydroxide ion by Lys261. DaCGO1 is specific for the 6-C-glucosyl flavones isovitexin, isoorientin and the 2-C-glucosyl xanthonoid mangiferin, and the subsequent C-C-bond cleavage by DaCGD generated apigenin, luteolin and norathyriol, respectively. Of the substrates tested, isovitexin was the preferred substrate (DaCGO1, K m 0.047 mM, k cat 51 min -1 ; DaCGO1/DaCGD, K m 0.083 mM, k cat 0.42 min -1 ).
Organizational Affiliation:
School of Engineering Sciences in Chemistry, Biotechnology, and Health, CBH, KTH Royal Institute of Technology, 100 44 Stockholm, Sweden.