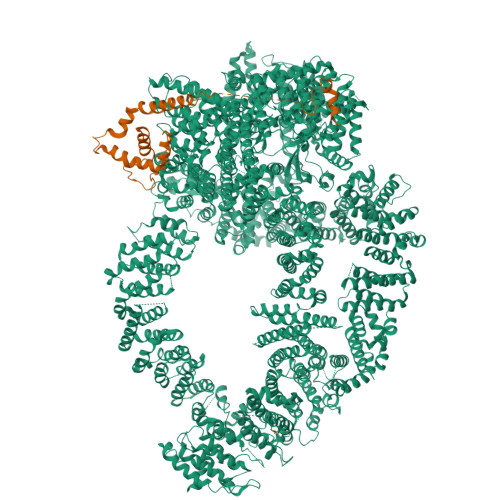
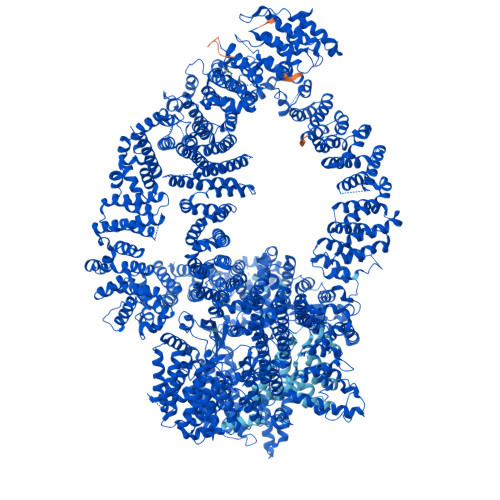
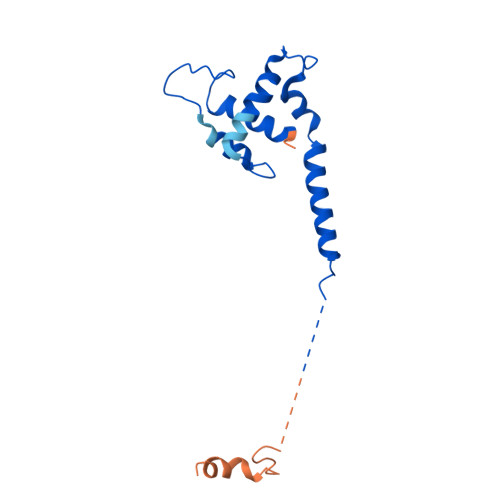
Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Li, C., Smirnova, E., Schnitzler, C., Crucifix, C., Concordet, J.P., Brion, A., Poterszman, A., Schultz, P., Papai, G., Ben-Shem, A.(2024) Nature 635: 764-769
- PubMed: 39260417
- DOI: https://doi.org/10.1038/s41586-024-08011-w
- Primary Citation of Related Structures:
8QR1, 8QRI - PubMed Abstract:
Chromatin structure is a key regulator of DNA transcription, replication, and repair 1 . In humans, the TIP60/EP400 complex (TIP60-C) is a 20-subunit assembly that impacts chromatin structure via two enzymatic activities: ATP-dependent exchange of histone H2A/H2B for H2A.Z/H2B and histone acetylation, which in yeast are carried out by two independent complexes, SWR1 and NuA4, respectively 2,3 . How these activities are merged in humans into one super-complex and what this association entails for their structure, mechanism and recruitment to chromatin is unknown. Here we describe the 2.4-3.3 Å resolution structure of the endogenous human TIP60-C. We find a three lobed architecture composed of SWR1-like (SWR1L) and NuA4-like (NuA4L) parts, that associate with a TRRAP activator-binding module. The huge EP400 subunit harbors the ATPase motor, traverses twice the junction between SWR1L and NuA4L, and constitutes the scaffold of the three-lobed architecture. NuA4L is completely re-arranged compared to its yeast counterpart. TRRAP is flexibly tethered to NuA4L, in stark contrast to its robust connection to the complete opposite side of yeast NuA4 4-7 . A modeled nucleosome bound to SWR1L, supported by activity tests, suggests that some aspects of the histone exchange mechanism diverge from the yeast example 8,9 . Furthermore, a fixed actin module, as opposed to the mobile actin subcomplex in SWR1 8 , the flexibility of TRRAP and the weak effect of extra-nucleosomal DNA on exchange activity, lead to a different, activator-based, mode of enlisting TIP60-C to chromatin.
Organizational Affiliation:
Université de Strasbourg, IGBMC UMR 7104 UMR S 1258, Illkirch, France.