Transition transferases prime bacterial capsule polymerization.
Litschko, C., Di Domenico, V., Schulze, J., Li, S., Ovchinnikova, O.G., Voskuilen, T., Bethe, A., Cifuente, J.O., Marina, A., Budde, I., Mast, T.A., Sulewska, M., Berger, M., Buettner, F.F.R., Lowary, T.L., Whitfield, C., Codee, J.D.C., Schubert, M., Guerin, M.E., Fiebig, T.(2025) Nat Chem Biol 21: 120-130
- PubMed: 38951648
- DOI: https://doi.org/10.1038/s41589-024-01664-8
- Primary Citation of Related Structures:
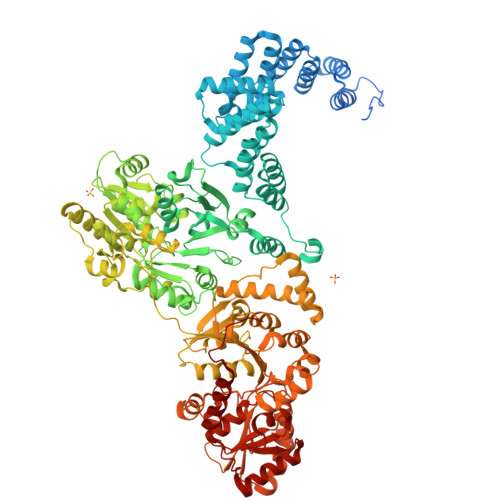
8QOY - PubMed Abstract:
Capsules are long-chain carbohydrate polymers that envelop the surfaces of many bacteria, protecting them from host immune responses. Capsule biosynthesis enzymes are potential drug targets and valuable biotechnological tools for generating vaccine antigens. Despite their importance, it remains unknown how structurally variable capsule polymers of Gram-negative pathogens are linked to the conserved glycolipid anchoring these virulence factors to the bacterial membrane. Using Actinobacillus pleuropneumoniae as an example, we demonstrate that CpsA and CpsC generate a poly(glycerol-3-phosphate) linker to connect the glycolipid with capsules containing poly(galactosylglycerol-phosphate) backbones. We reconstruct the entire capsule biosynthesis pathway in A. pleuropneumoniae serotypes 3 and 7, solve the X-ray crystal structure of the capsule polymerase CpsD, identify its tetratricopeptide repeat domain as essential for elongating poly(glycerol-3-phosphate) and show that CpsA and CpsC stimulate CpsD to produce longer polymers. We identify the CpsA and CpsC product as a wall teichoic acid homolog, demonstrating similarity between the biosynthesis of Gram-positive wall teichoic acid and Gram-negative capsules.
Organizational Affiliation:
Institute of Clinical Biochemistry, Hannover Medical School, Hannover, Germany.