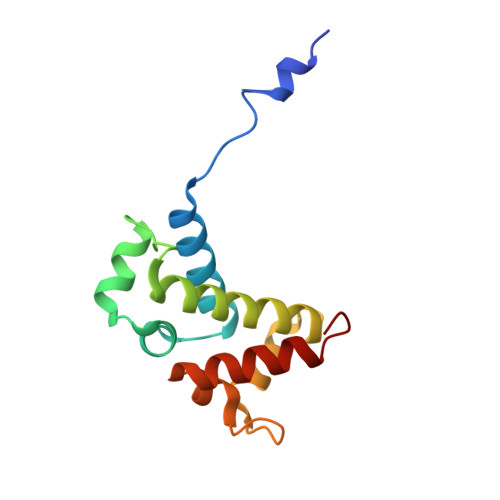
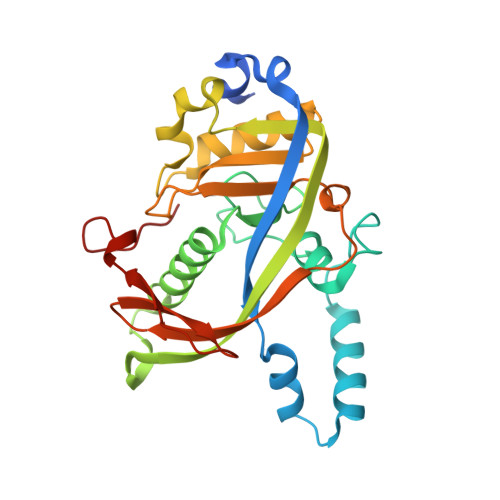
Toxin-mediated depletion of NAD and NADP drives persister formation in a human pathogen.
Santi, I., Dias Teixeira, R., Manfredi, P., Hernandez Gonzalez, H., Spiess, D.C., Mas, G., Klotz, A., Kaczmarczyk, A., Zamboni, N., Hiller, S., Jenal, U.(2024) EMBO J
- PubMed: 39322758
- DOI: https://doi.org/10.1038/s44318-024-00248-5
- Primary Citation of Related Structures:
8QNL, 8QNQ - PubMed Abstract:
Toxin-antitoxin (TA) systems are widespread in bacteria and implicated in genome stability, virulence, phage defense, and persistence. TA systems have diverse activities and cellular targets, but their physiological roles and regulatory mechanisms are often unclear. Here, we show that the NatR-NatT TA system, which is part of the core genome of the human pathogen Pseudomonas aeruginosa, generates drug-tolerant persisters by specifically depleting nicotinamide dinucleotides. While actively growing P. aeruginosa cells compensate for NatT-mediated NAD + deficiency by inducing the NAD + salvage pathway, NAD depletion generates drug-tolerant persisters under nutrient-limited conditions. Our structural and biochemical analyses propose a model for NatT toxin activation and autoregulation and indicate that NatT activity is subject to powerful metabolic feedback control by the NAD + precursor nicotinamide. Based on the identification of natT gain-of-function alleles in patient isolates and on the observation that NatT increases P. aeruginosa virulence, we postulate that NatT modulates pathogen fitness during infections. These findings pave the way for detailed investigations into how a toxin-antitoxin system can promote pathogen persistence by disrupting essential metabolic pathways.
Organizational Affiliation:
Biozentrum, University of Basel, Basel, Switzerland.