Structural Insights into the Activation of Human Aryl Hydrocarbon Receptor by the Environmental Contaminant Benzo[a]pyrene and Structurally Related Compounds.
Kwong, H.S., Paloni, M., Grandvuillemin, L., Sirounian, S., Ancelin, A., Lai-Kee-Him, J., Grimaldi, M., Carivenc, C., Lancey, C., Ragan, T.J., Hesketh, E.L., Balaguer, P., Barducci, A., Gruszczyk, J., Bourguet, W.(2024) J Mol Biol 436: 168411-168411
- PubMed: 38135181
- DOI: https://doi.org/10.1016/j.jmb.2023.168411
- Primary Citation of Related Structures:
8QMO - PubMed Abstract:
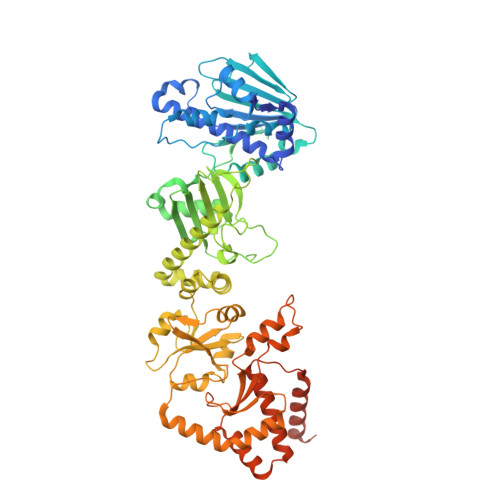
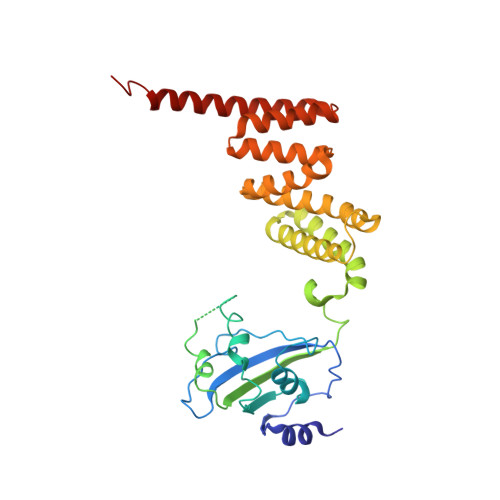
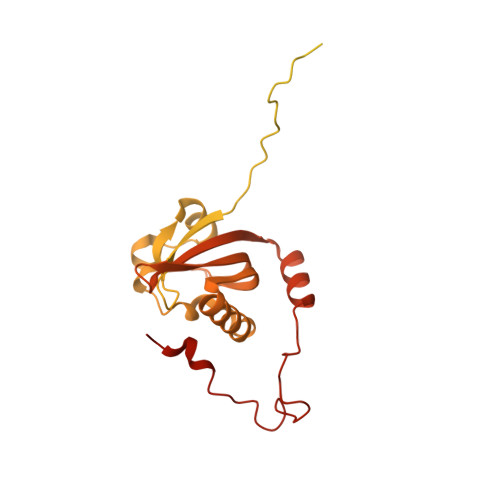
The aryl hydrocarbon receptor (AHR) is a ligand-dependent transcription factor belonging to the bHLH/PAS protein family and responding to hundreds of natural and chemical substances. It is primarily involved in the defense against chemical insults and bacterial infections or in the adaptive immune response, but also in the development of pathological conditions ranging from inflammatory to neoplastic disorders. Despite its prominent roles in many (patho)physiological processes, the lack of high-resolution structural data has precluded for thirty years an in-depth understanding of the structural mechanisms underlying ligand-binding specificity, promiscuity and activation of AHR. We recently reported a cryogenic electron microscopy (cryo-EM) structure of human AHR bound to the natural ligand indirubin, the chaperone Hsp90 and the co-chaperone XAP2 that provided the first experimental visualization of its ligand-binding PAS-B domain. Here, we report a 2.75 Å resolution structure of the AHR complex bound to the environmental pollutant benzo[a]pyrene (B[a]P). The structure substantiates the existence of a bipartite PAS-B ligand-binding pocket with a geometrically constrained primary binding site controlling ligand binding specificity and affinity, and a secondary binding site contributing to the binding promiscuity of AHR. We also report a docking study of B[a]P congeners that validates the B[a]P-bound PAS-B structure as a suitable model for accurate computational ligand binding assessment. Finally, comparison of our agonist-bound complex with the recently reported structures of mouse and fruit fly AHR PAS-B in different activation states suggests a ligand-induced loop conformational change potentially involved in the regulation of AHR function.
Organizational Affiliation:
CBS (Centre de Biologie Structurale), Univ Montpellier, CNRS, Inserm, Montpellier, France.