Pan-neutralizing antibody isolated from a COVID patient
Planchais, C., Fernandez, I., Rey, F.A., Mouquet, H.To be published.
Experimental Data Snapshot
Starting Models: experimental, in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Spike glycoprotein | A [auth E] | 243 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 | 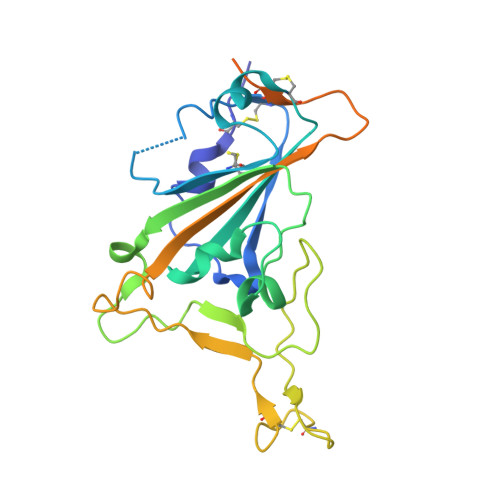 |
UniProt | |||||
Find proteins for A0A8A5XRG7 (Severe acute respiratory syndrome coronavirus 2) Explore A0A8A5XRG7 Go to UniProtKB: A0A8A5XRG7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8A5XRG7 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cv2.3194 heavy chain | B [auth H] | 229 | Homo sapiens | Mutation(s): 0 | 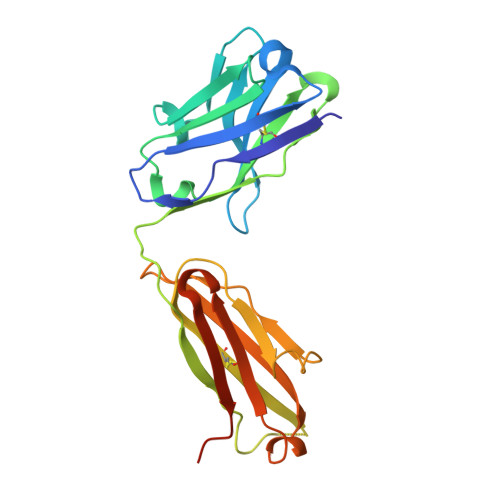 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| IGK@ protein | C [auth L] | 211 | Homo sapiens | Mutation(s): 0 Gene Names: IGK@ | 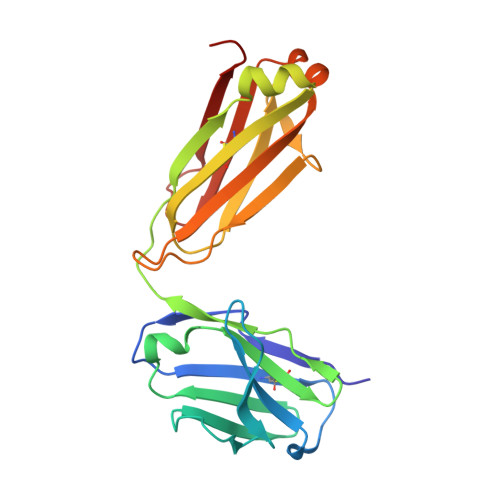 |
UniProt | |||||
Find proteins for Q6PJF2 (Homo sapiens) Explore Q6PJF2 Go to UniProtKB: Q6PJF2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6PJF2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | D [auth E] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57.793 | α = 90 |
| b = 85.348 | β = 97.48 |
| c = 193.987 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |