Characterization and structural study of a novel beta-N-acetylgalactosaminidase from Niabella aurantiaca.
Moreno Prieto, E.S., Fjermedal, S., Siebenhaar, S., Vuillemin, M., Holck, J., Vincentelli, R., Gippert, G.P., Wilkens, C., Morth, J.P., Henrissat, B.(2024) FEBS J 291: 1439-1456
- PubMed: 38129294
- DOI: https://doi.org/10.1111/febs.17042
- Primary Citation of Related Structures:
8P4L, 8Q8H - PubMed Abstract:
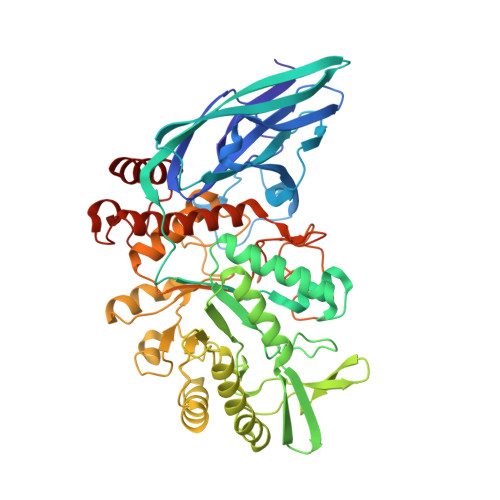
We report here the identification, characterization and three-dimensional (3D) structure determination of NaNga, a newly identified β-N-acetylgalactosaminidase from the Gram-negative soil bacterium Niabella aurantiaca DSM 17617. When recombinantly expressed in Escherichia coli, the enzyme selectively cleaved 4-nitrophenyl-N-acetyl-β-d-galactosamine (pNP-β-d-GalpNAc). The X-ray crystal structure of the protein was refined to 2.5 Å and consists of an N-terminal β-sandwich domain and a (β/α) 8 barrel catalytic domain. Despite a mere 22% sequence identity, the 3D structure of NaNga is similar to those previously determined for family GH123 members, suggesting it also employs the same substrate-assisted catalytic mechanism. Inhibition by N-acetyl-galactosamine thiazoline (GalNAc-thiazoline) supports the suggested mechanism. A phylogenetic analysis of its proximal sequence space shows significant clustering of unknown sequences around NaNga with sufficient divergence with previously identified GH123 members to subdivide this family into distinct subfamilies. Although the actual biological substrate of our enzyme remains unknown, examination of the active site pocket suggests that it may be a β-N-acetylgalactosaminide substituted by a monosaccharide at O-3. Analysis of the genomic context suggests, in turn, that this substituted β-N-acetylgalactosaminide may be appended to a d-arabinan from an environmental Actinomycete.
Organizational Affiliation:
Department of Biotechnology and Biomedicine, Technical University of Denmark, Kgs. Lyngby, Denmark.