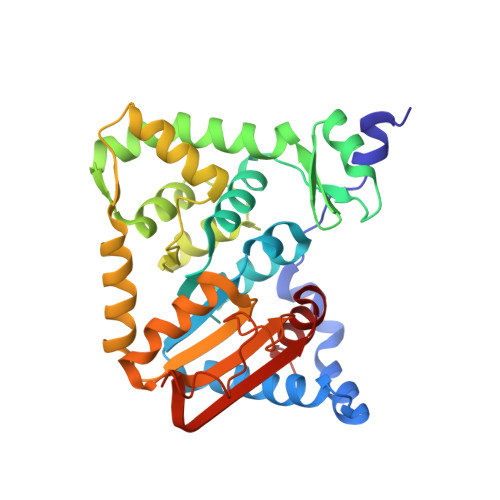
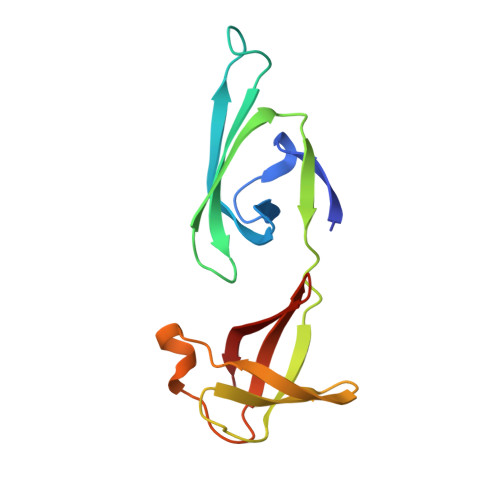
Crystal structure of archaeal IF5A-DHS complex reveals insights into the hypusination mechanism.
D'Agostino, M., Simonetti, A., Motta, S., Wolff, P., Romagnoli, A., Piccinini, A., Spinozzi, F., Di Marino, D., La Teana, A., Ennifar, E.(2024) Structure 32: 878
- PubMed: 38582076
- DOI: https://doi.org/10.1016/j.str.2024.03.008
- Primary Citation of Related Structures:
8PUT - PubMed Abstract:
The translation factor IF5A is highly conserved in Eukarya and Archaea and undergoes a unique post-translational hypusine modification by the deoxyhypusine synthase (DHS) enzyme. DHS transfers the butylamine moiety from spermidine to IF5A using NAD as a cofactor, forming a deoxyhypusine intermediate. IF5A is a key player in protein synthesis, preventing ribosome stalling in proline-rich sequences during translation elongation and facilitating translation elongation and termination. Additionally, human eIF5A participates in various essential cellular processes and contributes to cancer metastasis, with inhibiting hypusination showing anti-proliferative effects. The hypusination pathway of IF5A is therefore an attractive new therapeutic target. We elucidated the 2.0 Å X-ray crystal structure of the archaeal DHS-IF5A complex, revealing hetero-octameric architecture and providing a detailed view of the complex active site including the hypusination loop. This structure, along with biophysical data and molecular dynamics simulations, provides new insights into the catalytic mechanism of the hypusination reaction.
Organizational Affiliation:
Department of Life and Environmental Sciences, Polytechnic University of Marche, Via Brecce Bianche, 60131 Ancona, Italy; Architecture et Réactivité de l'ARN, CNRS UPR 9002, Institut de Biologie Moléculaire et Cellulaire, Université de Strasbourg, Strasbourg, France.