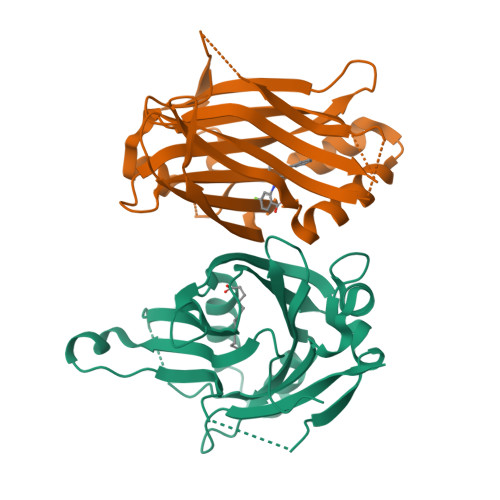
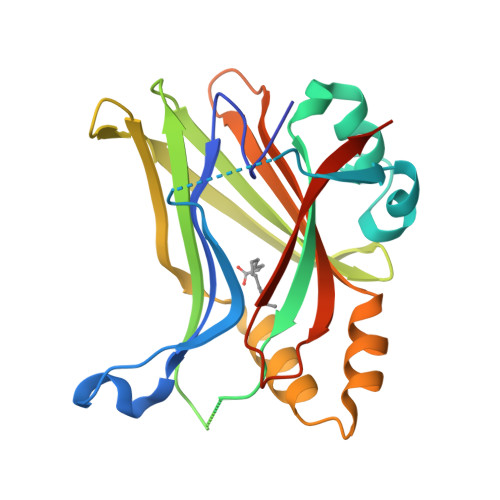
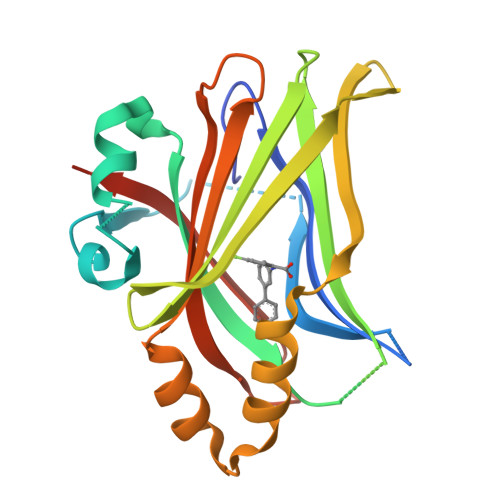
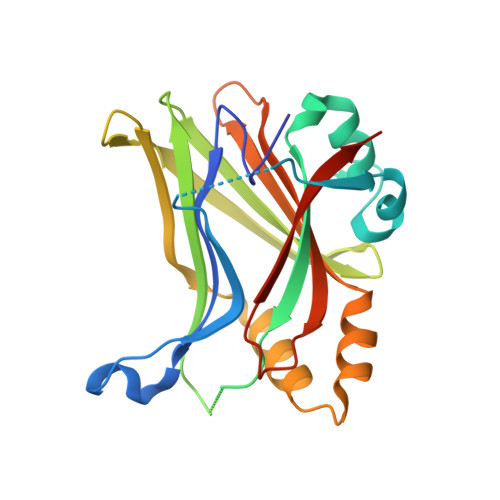
Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Fnaiche, A., Melin, L., Suarez, N.G., Paquin, A., Vu, V., Li, F., Allali-Hassani, A., Bolotokova, A., Allemand, F., Gelin, M., Cotelle, P., Woo, S., LaPlante, S.R., Barsyte-Lovejoy, D., Santhakumar, V., Vedadi, M., Guichou, J.F., Annabi, B., Gagnon, A.(2023) Bioorg Med Chem Lett 95: 129488-129488
- PubMed: 37770003
- DOI: https://doi.org/10.1016/j.bmcl.2023.129488
- Primary Citation of Related Structures:
8P29, 8POJ, 8POM, 8PON - PubMed Abstract:
The Hippo pathway regulates organ size and tissue homeostasis by controlling cell proliferation and apoptosis. The YAP-TEAD transcription factor, the downstream effector of the Hippo pathway, regulates the expression of genes such as CTGF, Cyr61, Axl and NF2. Aberrant Hippo activity has been identified in multiple types of cancers. Flufenamic acid (FA) was reported to bind in a liphophilic TEAD palmitic acid (PA) pocket, leading to reduction of the expression of Axl and NF2. Here, we show that the replacement of the trifluoromethyl moiety in FA by aromatic groups, directly connected to the scaffold or separated by a linker, leads to compounds with better affinity to TEAD. Co-crystallization studies show that these compounds bind similarly to FA, but deeper within the PA pocket. Our studies identified LM-41 and AF-2112 as two TEAD binders that strongly reduce the expression of CTGF, Cyr61, Axl and NF2. LM-41 gave the strongest reduction of migration of human MDA-MB-231 breast cancer cells.
Organizational Affiliation:
Département de chimie, Université du Québec à Montréal, C.P. 8888, Succursale Centre-Ville, Montréal, Québec H3C 3P8, Canada.