Structural analysis of chorismate mutase and cyclohexadienyl dehydratase from Pseudomonas aeruginosa
Khatanbaatar, T., Cordara, G., Krengel, U.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| chorismate mutase | 424 | Aequoribacter fuscus | Mutation(s): 0 Gene Names: EYZ66_08225 | 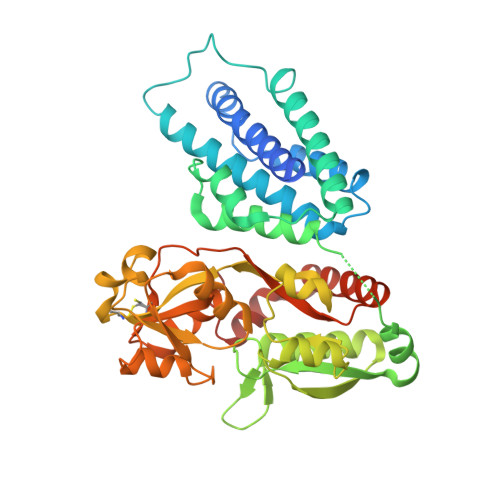 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| TSA (Subject of Investigation/LOI) Query on TSA | B [auth A] | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID C10 H12 O6 KRZHNRULRHECRF-JQCUSGDOSA-N |  | ||
| PEG (Subject of Investigation/LOI) Query on PEG | C [auth A], D [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL (Subject of Investigation/LOI) Query on GOL | E [auth A], F [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL (Subject of Investigation/LOI) Query on CL | G [auth A], H [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 48.608 | α = 90 |
| b = 105.344 | β = 112.484 |
| c = 55.776 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | 310030M_182648 |