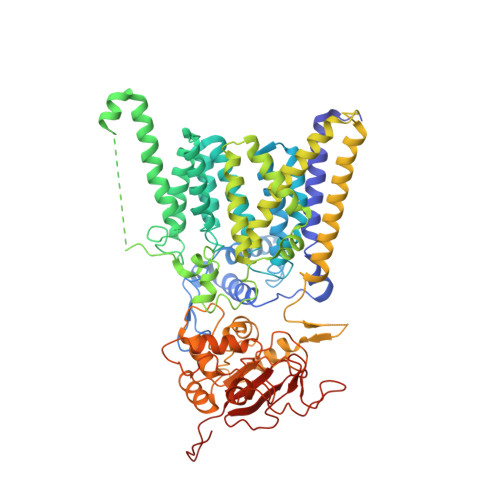
Positive selection CRISPR screens reveal a druggable pocket in an oligosaccharyltransferase required for inflammatory signaling to NF-kappa B.
Lampson, B.L., Ramirez, A.S., Baro, M., He, L., Hegde, M., Koduri, V., Pfaff, J.L., Hanna, R.E., Kowal, J., Shirole, N.H., He, Y., Doench, J.G., Contessa, J.N., Locher, K.P., Kaelin Jr., W.G.(2024) Cell 187: 2209-2223.e16
- PubMed: 38670073
- DOI: https://doi.org/10.1016/j.cell.2024.03.022
- Primary Citation of Related Structures:
8PN9 - PubMed Abstract:
Nuclear factor κB (NF-κB) plays roles in various diseases. Many inflammatory signals, such as circulating lipopolysaccharides (LPSs), activate NF-κB via specific receptors. Using whole-genome CRISPR-Cas9 screens of LPS-treated cells that express an NF-κB-driven suicide gene, we discovered that the LPS receptor Toll-like receptor 4 (TLR4) is specifically dependent on the oligosaccharyltransferase complex OST-A for N-glycosylation and cell-surface localization. The tool compound NGI-1 inhibits OST complexes in vivo, but the underlying molecular mechanism remained unknown. We did a CRISPR base-editor screen for NGI-1-resistant variants of STT3A, the catalytic subunit of OST-A. These variants, in conjunction with cryoelectron microscopy studies, revealed that NGI-1 binds the catalytic site of STT3A, where it traps a molecule of the donor substrate dolichyl-PP-GlcNAc 2 -Man 9 -Glc 3 , suggesting an uncompetitive inhibition mechanism. Our results provide a rationale for and an initial step toward the development of STT3A-specific inhibitors and illustrate the power of contemporaneous base-editor and structural studies to define drug mechanism of action.
Organizational Affiliation:
Department of Medical Oncology, Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA 02215, USA.