Octahedrally coordinated iron in the catalytic site of endonuclease IV from Staphylococcus aureus
Kirillov, S., Isupov, M.N., Paterson, N., Wiener, R., Abeldenov, S., Saper, M.A., Rouvinski, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Probable endonuclease 4 | 296 | Staphylococcus aureus | Mutation(s): 0 Gene Names: nfo, SAR1634 EC: 3.1.21.2 | 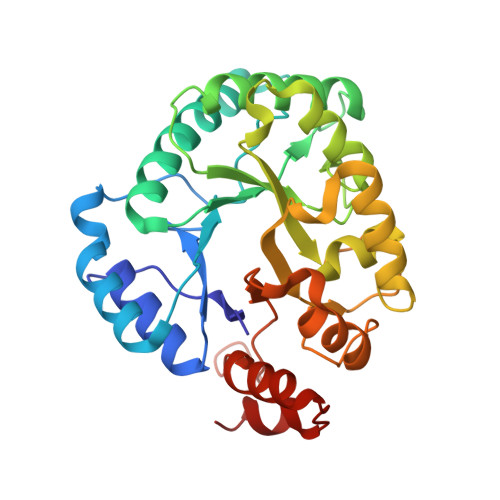 | |
UniProt | |||||
Find proteins for Q6GGE2 (Staphylococcus aureus (strain MRSA252)) Explore Q6GGE2 Go to UniProtKB: Q6GGE2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6GGE2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PO4 (Subject of Investigation/LOI) Query on PO4 | E [auth A] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | D [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| FE (Subject of Investigation/LOI) Query on FE | B [auth A], C [auth A] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A], G [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 38.548 | α = 90 |
| b = 84.04 | β = 90 |
| c = 103.344 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | -- |