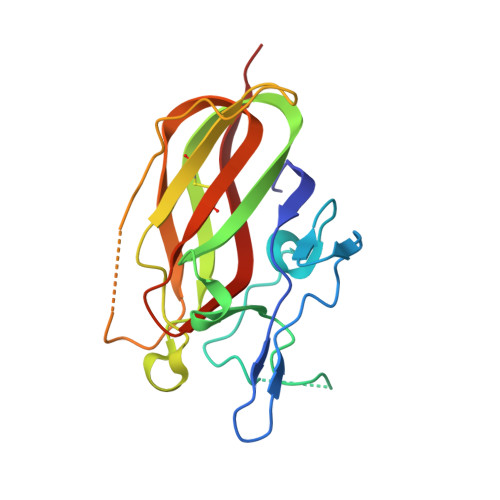
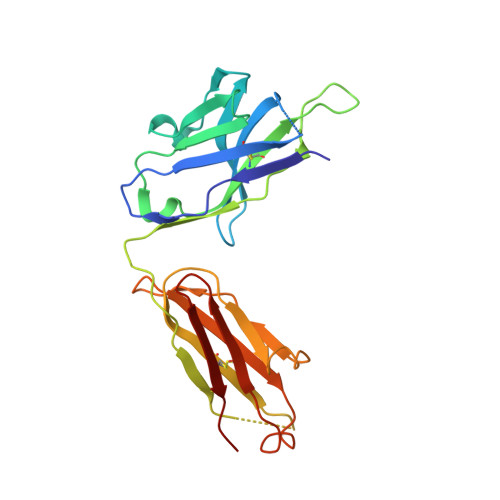
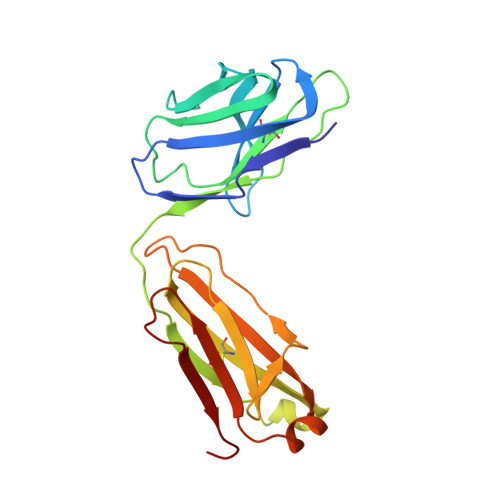
A highly selective humanized DDR1 mAb reverses immune exclusion by disrupting collagen fiber alignment in breast cancer.
Liu, J., Chiang, H.C., Xiong, W., Laurent, V., Griffiths, S.C., Dulfer, J., Deng, H., Sun, X., Yin, Y.W., Li, W., Audoly, L.P., An, Z., Schurpf, T., Li, R., Zhang, N.(2023) J Immunother Cancer 11
- PubMed: 37328286
- DOI: https://doi.org/10.1136/jitc-2023-006720
- Primary Citation of Related Structures:
8PE9 - PubMed Abstract:
Immune exclusion (IE) where tumors deter the infiltration of immune cells into the tumor microenvironment has emerged as a key mechanism underlying immunotherapy resistance. We recently reported a novel role of discoidin domain-containing receptor 1 (DDR1) in promoting IE in breast cancer and validated its critical role in IE using neutralizing rabbit monoclonal antibodies (mAbs) in multiple mouse tumor models. To develop a DDR1-targeting mAb as a potential cancer therapeutic, we humanized mAb9 with a complementarity-determining region grafting strategy. The humanized antibody named PRTH-101 is currently being tested in a Phase 1 clinical trial. We determined the binding epitope of PRTH-101 from the crystal structure of the complex between DDR1 extracellular domain (ECD) and the PRTH-101 Fab fragment with 3.15 Å resolution. We revealed the underlying mechanisms of action of PRTH-101 using both cell culture assays and in vivo study in a mouse tumor model. PRTH-101 has subnanomolar affinity to DDR1 and potent antitumor efficacy similar to the parental rabbit mAb after humanization. Structural information illustrated that PRTH-101 interacts with the discoidin (DS)-like domain, but not the collagen-binding DS domain of DDR1. Mechanistically, we showed that PRTH-101 inhibited DDR1 phosphorylation, decreased collagen-mediated cell attachment, and significantly blocked DDR1 shedding from the cell surface. Treatment of tumor-bearing mice with PRTH-101 in vivo disrupted collagen fiber alignment (a physical barrier) in the tumor extracellular matrix (ECM) and enhanced CD8 + T cell infiltration in tumors. This study not only paves a pathway for the development of PRTH-101 as a cancer therapeutic, but also sheds light on a new therapeutic strategy to modulate collagen alignment in the tumor ECM for enhancing antitumor immunity.
Organizational Affiliation:
Texas Therapeutics Institute, Brown Foundation Institute of Molecular Medicine, The University of Texas Health Science Center at Houston, Houston, Texas, USA.