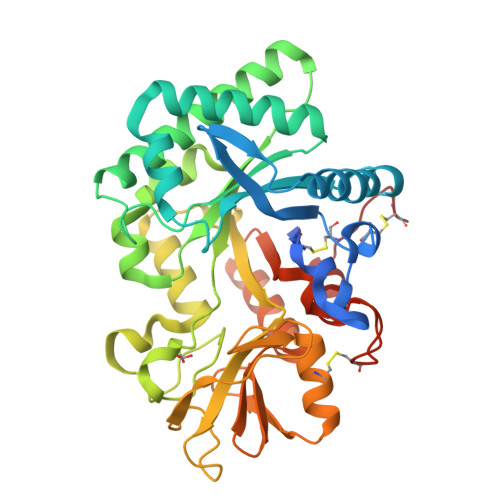
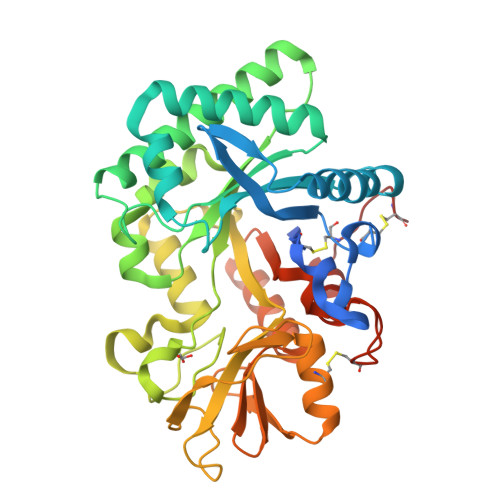
Ym1 protein crystals promote type 2 immunity.
Heyndrickx, I., Deswarte, K., Verstraete, K., Verschueren, K.H.G., Smole, U., Aegerter, H., Dansercoer, A., Hammad, H., Savvides, S.N., Lambrecht, B.N.(2024) Elife 12
- PubMed: 38194250
- DOI: https://doi.org/10.7554/eLife.90676
- Primary Citation of Related Structures:
8P8Q, 8P8R, 8P8S, 8P8T - PubMed Abstract:
Spontaneous protein crystallization is a rare event, yet protein crystals are frequently found in eosinophil-rich inflammation. In humans, Charcot-Leyden crystals (CLCs) are made from galectin-10 (Gal10) protein, an abundant protein in eosinophils. Although mice do not encode Gal10 in their genome, they do form pseudo-CLCs, made from the chitinase-like proteins Ym1 and/or Ym2, encoded by Chil3 and Chil4 and made by myeloid and epithelial cells respectively. Here, we investigated the biological effects of pseudo-CLCs since their function is currently unknown. We produced recombinant Ym1 crystals which were shown to have identical crystal packing and structure by X-ray crystallography as in vivo native crystals derived from murine lung. When administered to the airways of mice, crystalline but not soluble Ym1 stimulated innate and adaptive immunity and acted as a type 2 immune adjuvant for eosinophilic inflammation via triggering of dendritic cells (DCs). Murine Ym1 protein crystals found at sites of eosinophilic inflammation reinforce type 2 immunity and could serve as a surrogate model for studying the biology of human CLCs.
Organizational Affiliation:
Laboratory of Immunoregulation and Mucosal Immunology, VIB-UGent Center for Inflammation Research, Ghent, Belgium.