Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Wang, G., Seidler, N.J., Rohm, S., Pan, Y., Liang, X.J., Haarer, L., Berger, B.T., Sivashanmugam, S.A., Wydra, V.R., Forster, M., Laufer, S.A., Chaikuad, A., Gehringer, M., Knapp, S.(2024) Angew Chem Int Ed Engl : e202419736-e202419736
- PubMed: 39716901
- DOI: https://doi.org/10.1002/anie.202419736
- Primary Citation of Related Structures:
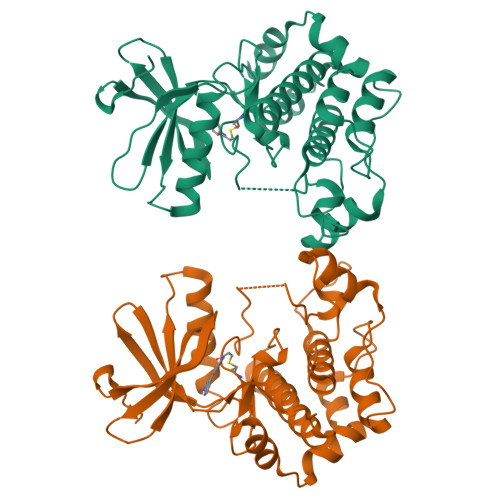
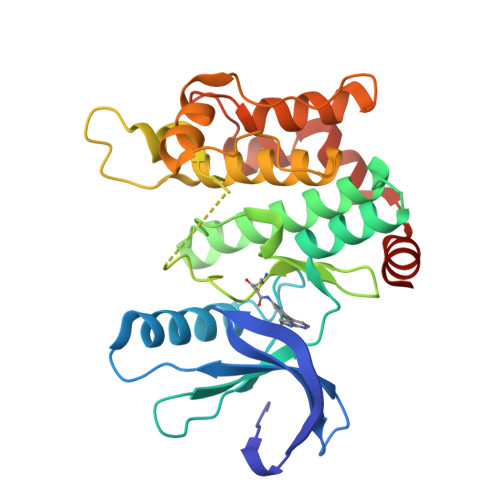
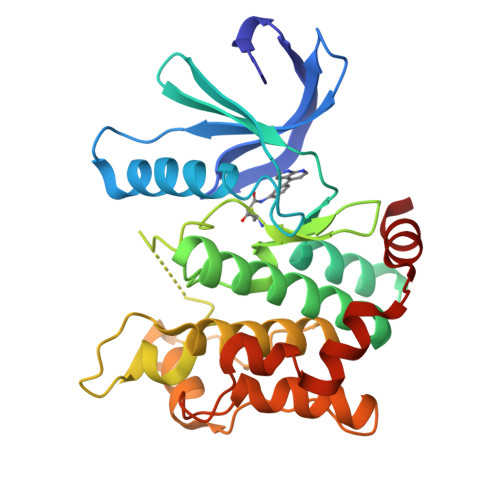
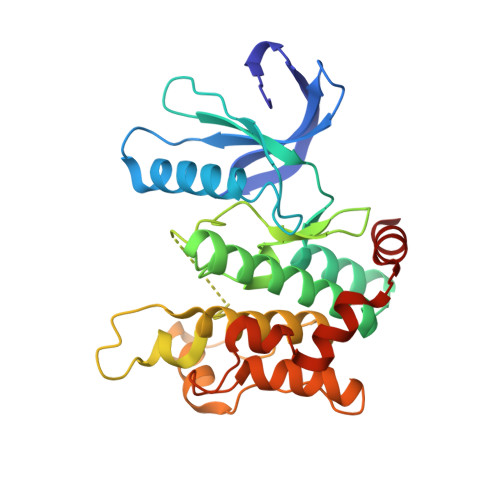
8P7J, 8PM3, 9F32, 9F81, 9HHW - PubMed Abstract:
Protein kinases are important drug targets, yet specific inhibitors have been developed for only a fraction of the more than 500 human kinases. A major challenge in designing inhibitors for highly related kinases is selectivity. Unlike their non-covalent counterparts, covalent inhibitors offer the advantage of selectively targeting structurally similar kinases by modifying specific protein side chains, particularly non-conserved cysteines. Previously, covalent fragment screens yielded potent and selective compounds for individual kinases such as ERK1/2 but have not been applied to the broader kinome. Furthermore, many of the accessible cysteine positions have not been addressed so far. Here, we outline a generalizable approach to sample ATP-site cysteines with fragment-like covalent inhibitors. We present the development of a kinase-focused fragment library and its systematic screening against a curated selection of 47 kinases, with 60 active site-proximal cysteines using LC/MS and differential scanning fluorimetry (DSF) assays, followed by hit validation through various complementary techniques. Our findings expand the repertoire of targetable cysteines within protein kinases, provide insight into unique binding modes identified from crystal structures and deliver isoform-specific hits with promising profiles as starting points for the development of highly potent and selective covalent inhibitors.
Organizational Affiliation:
Goethe-Universitat Frankfurt am Main Fachbereich 14 Biochemie Chemie und Pharmazie, Institute for Pharmaceutical Chemistry, GERMANY.