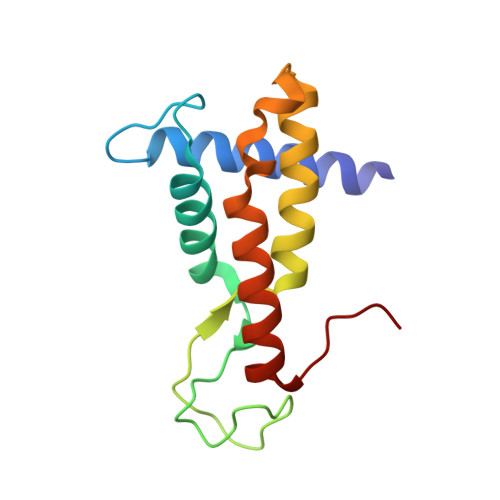
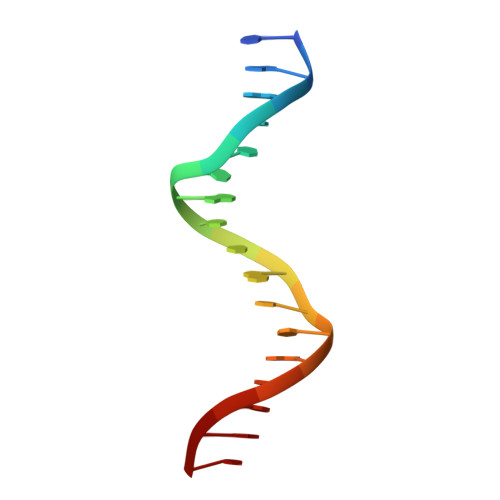
The ALOG domain defines a family of plant-specific transcription factors acting during Arabidopsis flower development.
Rieu, P., Beretta, V.M., Caselli, F., Thevenon, E., Lucas, J., Rizk, M., Franchini, E., Caporali, E., Paleni, C., Nanao, M.H., Kater, M.M., Dumas, R., Zubieta, C., Parcy, F., Gregis, V.(2024) Proc Natl Acad Sci U S A 121: e2310464121-e2310464121
- PubMed: 38412122
- DOI: https://doi.org/10.1073/pnas.2310464121
- Primary Citation of Related Structures:
8P5Q - PubMed Abstract:
The ALOG (Arabidopsis LIGHT-DEPENDENT SHORT HYPOCOTYLS 1 (LSH1) and Oryza G1) proteins are conserved plant-specific Transcription Factors (TFs). They play critical roles in the development of various plant organs (meristems, inflorescences, floral organs, and nodules) from bryophytes to higher flowering plants. Despite the fact that the first members of this family were originally discovered in Arabidopsis, their role in this model plant has remained poorly characterized. Moreover, how these transcriptional regulators work at the molecular level is unknown. Here, we study the redundant function of the ALOG proteins LSH1,3,4 from Arabidopsis. We uncover their role in the repression of bract development and position them within a gene regulatory network controlling this process and involving the floral regulators LEAFY, BLADE-ON-PETIOLE, and PUCHI. Next, using in vitro genome-wide studies, we identified the conserved DNA motif bound by ALOG proteins from evolutionarily distant species (the liverwort Marchantia polymorpha and the flowering plants Arabidopsis, tomato, and rice). Resolution of the crystallographic structure of the ALOG DNA-binding domain in complex with DNA revealed the domain is a four-helix bundle with a disordered NLS and a zinc ribbon insertion between helices 2 and 3. The majority of DNA interactions are mediated by specific contacts made by the third alpha helix and the NLS. Taken together, this work provides the biochemical and structural basis for DNA-binding specificity of an evolutionarily conserved TF family and reveals its role as a key player in Arabidopsis flower development.
Organizational Affiliation:
Laboratoire Physiologie Cellulaire et Végétale, Université Grenoble Alpes, Centre national de la recherche scientifique, Commissariat à l'énergie atomique et aux énergies alternatives, Institut national de recherche pour l'agriculture, l'alimentation et l'environnement, Département de Biologie Structurale et Cellulaire intégrée, Grenoble F-38054, France.