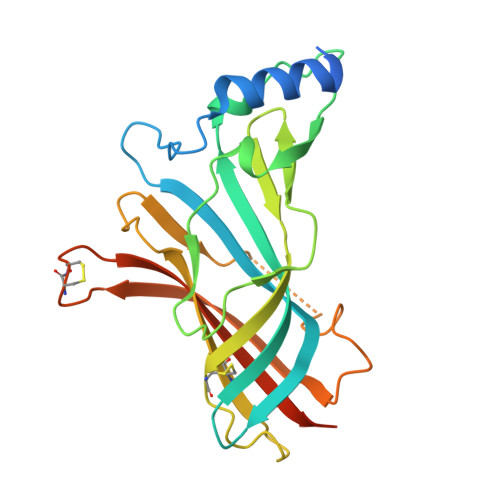
Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
Fitzgerald, E.A., Cederfelt, D., Boronat, P., Aarmo Lund, B., de Esch, I.J.P., Danielson, U.H.To be published.